Note
Click here to download the full example code
Running NiBetaSeries¶
This example runs through a basic call of NiBetaSeries using
the commandline entry point nibs.
While this example is using python, typically nibs will be
called directly on the commandline.
Import all the necessary packages¶
import tempfile # make a temporary directory for files
import os # interact with the filesystem
import urllib.request # grad data from internet
import tarfile # extract files from tar
from subprocess import Popen, PIPE, STDOUT # enable calling commandline
import matplotlib.pyplot as plt # manipulate figures
import seaborn as sns # display results
import pandas as pd # manipulate tabular data
Download relevant data from ds000164 (and Atlas Files)¶
The subject data came from openneuro [notebook-1]. The atlas data came from a recently published parcellation in a publically accessible github repository.
# atlas github repo for reference:
"""https://github.com/ThomasYeoLab/CBIG/raw/master/stable_projects/\
brain_parcellation/Schaefer2018_LocalGlobal/Parcellations/MNI/"""
data_dir = tempfile.mkdtemp()
print('Our working directory: {}'.format(data_dir))
# download the tar data
url = "https://www.dropbox.com/s/qoqbiya1ou7vi78/ds000164-test_v1.tar.gz?dl=1"
tar_file = os.path.join(data_dir, "ds000164.tar.gz")
u = urllib.request.urlopen(url)
data = u.read()
u.close()
# write tar data to file
with open(tar_file, "wb") as f:
f.write(data)
# extract the data
tar = tarfile.open(tar_file, mode='r|gz')
tar.extractall(path=data_dir)
os.remove(tar_file)
Out:
Our working directory: /tmp/tmpfobybnla
Display the minimal dataset necessary to run nibs¶
# https://stackoverflow.com/questions/9727673/list-directory-tree-structure-in-python
def list_files(startpath):
for root, dirs, files in os.walk(startpath):
level = root.replace(startpath, '').count(os.sep)
indent = ' ' * 4 * (level)
print('{}{}/'.format(indent, os.path.basename(root)))
subindent = ' ' * 4 * (level + 1)
for f in files:
print('{}{}'.format(subindent, f))
list_files(data_dir)
Out:
tmpfobybnla/
ds000164/
T1w.json
README
task-stroop_bold.json
dataset_description.json
task-stroop_events.json
CHANGES
derivatives/
data/
Schaefer2018_100Parcels_7Networks_order.txt
Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz
fmriprep/
sub-001/
func/
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_brainmask.nii.gz
sub-001_task-stroop_bold_confounds.tsv
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz
sub-001/
anat/
sub-001_T1w.nii.gz
func/
sub-001_task-stroop_bold.nii.gz
sub-001_task-stroop_events.tsv
Manipulate events file so it satifies assumptions¶
1. the correct column has 1’s and 0’s corresponding to correct and incorrect, respectively. 2. the condition column is renamed to trial_type nibs currently depends on the “correct” column being binary and the “trial_type” column to contain the trial types of interest.
read the file¶
events_file = os.path.join(data_dir,
"ds000164",
"sub-001",
"func",
"sub-001_task-stroop_events.tsv")
events_df = pd.read_csv(events_file, sep='\t', na_values="n/a")
print(events_df.head())
Out:
onset duration correct condition response_time
0 0.342 1 Y neutral 1.186
1 3.345 1 Y congruent 0.667
2 12.346 1 Y congruent 0.614
3 15.349 1 Y neutral 0.696
4 18.350 1 Y neutral 0.752
replace condition with trial_type¶
events_df.rename({"condition": "trial_type"}, axis='columns', inplace=True)
print(events_df.head())
Out:
onset duration correct trial_type response_time
0 0.342 1 Y neutral 1.186
1 3.345 1 Y congruent 0.667
2 12.346 1 Y congruent 0.614
3 15.349 1 Y neutral 0.696
4 18.350 1 Y neutral 0.752
save the file¶
events_df.to_csv(events_file, sep="\t", na_rep="n/a", index=False)
Manipulate the region order file¶
There are several adjustments to the atlas file that need to be completed before we can pass it into nibs. Importantly, the relevant column names MUST be named “index” and “regions”. “index” refers to which integer within the file corresponds to which region in the atlas nifti file. “regions” refers the name of each region in the atlas nifti file.
read the atlas file¶
atlas_txt = os.path.join(data_dir,
"ds000164",
"derivatives",
"data",
"Schaefer2018_100Parcels_7Networks_order.txt")
atlas_df = pd.read_csv(atlas_txt, sep="\t", header=None)
print(atlas_df.head())
Out:
0 1 2 3 4 5
0 1 7Networks_LH_Vis_1 120 18 131 0
1 2 7Networks_LH_Vis_2 120 18 132 0
2 3 7Networks_LH_Vis_3 120 18 133 0
3 4 7Networks_LH_Vis_4 120 18 135 0
4 5 7Networks_LH_Vis_5 120 18 136 0
drop coordinate columns¶
atlas_df.drop([2, 3, 4, 5], axis='columns', inplace=True)
print(atlas_df.head())
Out:
0 1
0 1 7Networks_LH_Vis_1
1 2 7Networks_LH_Vis_2
2 3 7Networks_LH_Vis_3
3 4 7Networks_LH_Vis_4
4 5 7Networks_LH_Vis_5
rename columns with the approved headings: “index” and “regions”¶
atlas_df.rename({0: 'index', 1: 'regions'}, axis='columns', inplace=True)
print(atlas_df.head())
Out:
index regions
0 1 7Networks_LH_Vis_1
1 2 7Networks_LH_Vis_2
2 3 7Networks_LH_Vis_3
3 4 7Networks_LH_Vis_4
4 5 7Networks_LH_Vis_5
remove prefix “7Networks”¶
atlas_df.replace(regex={'7Networks_(.*)': '\\1'}, inplace=True)
print(atlas_df.head())
Out:
index regions
0 1 LH_Vis_1
1 2 LH_Vis_2
2 3 LH_Vis_3
3 4 LH_Vis_4
4 5 LH_Vis_5
write out the file as .tsv¶
atlas_tsv = atlas_txt.replace(".txt", ".tsv")
atlas_df.to_csv(atlas_tsv, sep="\t", index=False)
Run nibs¶
out_dir = os.path.join(data_dir, "ds000164", "derivatives")
work_dir = os.path.join(out_dir, "work")
atlas_mni_file = os.path.join(data_dir,
"ds000164",
"derivatives",
"data",
"Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz")
cmd = """\
nibs -c WhiteMatter CSF \
--participant-label 001 \
-w {work_dir} \
-a {atlas_mni_file} \
-l {atlas_tsv} \
{bids_dir} \
fmriprep \
{out_dir} \
participant
""".format(atlas_mni_file=atlas_mni_file,
atlas_tsv=atlas_tsv,
bids_dir=os.path.join(data_dir, "ds000164"),
out_dir=out_dir,
work_dir=work_dir)
# Since we cannot run bash commands inside this tutorial
# we are printing the actual bash command so you can see it
# in the output
print("The Example Command:\n", cmd)
# call nibs
p = Popen(cmd, shell=True, stdout=PIPE, stderr=STDOUT)
while True:
line = p.stdout.readline()
if not line:
break
print(line)
Out:
The Example Command:
nibs -c WhiteMatter CSF --participant-label 001 -w /tmp/tmpfobybnla/ds000164/derivatives/work -a /tmp/tmpfobybnla/ds000164/derivatives/data/Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz -l /tmp/tmpfobybnla/ds000164/derivatives/data/Schaefer2018_100Parcels_7Networks_order.tsv /tmp/tmpfobybnla/ds000164 fmriprep /tmp/tmpfobybnla/ds000164/derivatives participant
b'190904-03:43:39,765 nipype.workflow INFO:\n'
b"\t Workflow nibetaseries_participant_wf settings: ['check', 'execution', 'logging', 'monitoring']\n"
b'190904-03:43:39,777 nipype.workflow INFO:\n'
b'\t Running in parallel.\n'
b'190904-03:43:39,780 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b"/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.3.1/lib/python3.7/site-packages/grabbit/core.py:449: UserWarning: Domain with name 'bids' already exists; returning existing Domain configuration.\n"
b' warnings.warn(msg)\n'
b'190904-03:43:39,813 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.betaseries_wf.betaseries_node" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/betaseries_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/betaseries_node".\n'
b'190904-03:43:39,817 nipype.workflow INFO:\n'
b'\t [Node] Running "betaseries_node" ("nibetaseries.interfaces.nistats.BetaSeries")\n'
b'190904-03:43:41,782 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 1 tasks, and 0 jobs ready. Free memory (GB): 6.81/7.01, Free processors: 3/4.\n'
b' Currently running:\n'
b' * nibetaseries_participant_wf.single_subject001_wf.betaseries_wf.betaseries_node\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.3.1/lib/python3.7/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 216, got 192\n'
b' return f(*args, **kwds)\n'
b"/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.3.1/lib/python3.7/importlib/_bootstrap.py:219: ImportWarning: can't resolve package from __spec__ or __package__, falling back on __name__ and __path__\n"
b' return f(*args, **kwds)\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b"<string>:6: DeprecationWarning: object of type <class 'numpy.float64'> cannot be safely interpreted as an integer.\n"
b"<string>:6: DeprecationWarning: object of type <class 'float'> cannot be safely interpreted as an integer.\n"
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'190904-03:44:56,524 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.betaseries_wf.betaseries_node".\n'
b'190904-03:44:57,858 nipype.workflow INFO:\n'
b'\t [Job 0] Completed (nibetaseries_participant_wf.single_subject001_wf.betaseries_wf.betaseries_node).\n'
b'190904-03:44:57,861 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190904-03:44:59,861 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190904-03:44:59,887 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node0" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/atlas_corr_node/mapflow/_atlas_corr_node0".\n'
b'190904-03:44:59,888 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node1" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/atlas_corr_node/mapflow/_atlas_corr_node1".\n'
b'190904-03:44:59,888 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node2" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/atlas_corr_node/mapflow/_atlas_corr_node2".\n'
b'190904-03:44:59,891 nipype.workflow INFO:\n'
b'\t [Node] Running "_atlas_corr_node0" ("nibetaseries.interfaces.nilearn.AtlasConnectivity")\n'
b'190904-03:44:59,892 nipype.workflow INFO:\n'
b'\t [Node] Running "_atlas_corr_node2" ("nibetaseries.interfaces.nilearn.AtlasConnectivity")\n'
b'190904-03:44:59,892 nipype.workflow INFO:\n'
b'\t [Node] Running "_atlas_corr_node1" ("nibetaseries.interfaces.nilearn.AtlasConnectivity")\n'
b'190904-03:45:01,863 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 3 tasks, and 0 jobs ready. Free memory (GB): 6.41/7.01, Free processors: 1/4.\n'
b' Currently running:\n'
b' * _atlas_corr_node2\n'
b' * _atlas_corr_node1\n'
b' * _atlas_corr_node0\n'
b'[NiftiLabelsMasker.fit_transform] loading data from /tmp/tmpfobybnla/ds000164/derivatives/data/Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz\n'
b'Resampling labels\n'
b'[NiftiLabelsMasker.transform_single_imgs] Loading data from /tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/betaseries_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/betaseries_node/betaseries_trialtyp\n'
b'[NiftiLabelsMasker.transform_single_imgs] Extracting region signals\n'
b'[NiftiLabelsMasker.transform_single_imgs] Cleaning extracted signals\n'
b'190904-03:45:09,185 nipype.workflow INFO:\n'
b'\t [Node] Finished "_atlas_corr_node1".\n'
b'[NiftiLabelsMasker.fit_transform] loading data from /tmp/tmpfobybnla/ds000164/derivatives/data/Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz\n'
b'Resampling labels\n'
b'[NiftiLabelsMasker.transform_single_imgs] Loading data from /tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/betaseries_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/betaseries_node/betaseries_trialtyp\n'
b'[NiftiLabelsMasker.transform_single_imgs] Extracting region signals\n'
b'[NiftiLabelsMasker.transform_single_imgs] Cleaning extracted signals\n'
b'190904-03:45:09,431 nipype.workflow INFO:\n'
b'\t [Node] Finished "_atlas_corr_node0".\n'
b'[NiftiLabelsMasker.fit_transform] loading data from /tmp/tmpfobybnla/ds000164/derivatives/data/Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz\n'
b'Resampling labels\n'
b'[NiftiLabelsMasker.transform_single_imgs] Loading data from /tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/betaseries_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/betaseries_node/betaseries_trialtyp\n'
b'[NiftiLabelsMasker.transform_single_imgs] Extracting region signals\n'
b'[NiftiLabelsMasker.transform_single_imgs] Cleaning extracted signals\n'
b'190904-03:45:09,674 nipype.workflow INFO:\n'
b'\t [Node] Finished "_atlas_corr_node2".\n'
b'190904-03:45:09,871 nipype.workflow INFO:\n'
b'\t [Job 5] Completed (_atlas_corr_node0).\n'
b'190904-03:45:09,872 nipype.workflow INFO:\n'
b'\t [Job 6] Completed (_atlas_corr_node1).\n'
b'190904-03:45:09,872 nipype.workflow INFO:\n'
b'\t [Job 7] Completed (_atlas_corr_node2).\n'
b'190904-03:45:09,873 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190904-03:45:09,895 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.correlation_wf.atlas_corr_node" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/atlas_corr_node".\n'
b'190904-03:45:09,898 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node0" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/atlas_corr_node/mapflow/_atlas_corr_node0".\n'
b'190904-03:45:09,899 nipype.workflow INFO:\n'
b'\t [Node] Cached "_atlas_corr_node0" - collecting precomputed outputs\n'
b'190904-03:45:09,899 nipype.workflow INFO:\n'
b'\t [Node] "_atlas_corr_node0" found cached.\n'
b'190904-03:45:09,900 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node1" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/atlas_corr_node/mapflow/_atlas_corr_node1".\n'
b'190904-03:45:09,901 nipype.workflow INFO:\n'
b'\t [Node] Cached "_atlas_corr_node1" - collecting precomputed outputs\n'
b'190904-03:45:09,901 nipype.workflow INFO:\n'
b'\t [Node] "_atlas_corr_node1" found cached.\n'
b'190904-03:45:09,901 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node2" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/atlas_corr_node/mapflow/_atlas_corr_node2".\n'
b'190904-03:45:09,902 nipype.workflow INFO:\n'
b'\t [Node] Cached "_atlas_corr_node2" - collecting precomputed outputs\n'
b'190904-03:45:09,902 nipype.workflow INFO:\n'
b'\t [Node] "_atlas_corr_node2" found cached.\n'
b'190904-03:45:09,904 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.correlation_wf.atlas_corr_node".\n'
b'190904-03:45:11,873 nipype.workflow INFO:\n'
b'\t [Job 1] Completed (nibetaseries_participant_wf.single_subject001_wf.correlation_wf.atlas_corr_node).\n'
b'190904-03:45:11,875 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 2 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190904-03:45:13,876 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 6 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190904-03:45:13,898 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_fig0" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/ds_correlation_fig/mapflow/_ds_correlation_fig0".\n'
b'190904-03:45:13,899 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_fig1" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/ds_correlation_fig/mapflow/_ds_correlation_fig1".\n'
b'190904-03:45:13,899 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_fig0" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190904-03:45:13,900 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_fig2" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/ds_correlation_fig/mapflow/_ds_correlation_fig2".\n'
b'190904-03:45:13,901 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_fig1" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190904-03:45:13,901 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_rename_matrix_node0" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/rename_matrix_node/mapflow/_rename_matrix_node0".\n'
b'190904-03:45:13,902 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_fig2" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190904-03:45:13,904 nipype.workflow INFO:\n'
b'\t [Node] Running "_rename_matrix_node0" ("nipype.interfaces.utility.wrappers.Function")\n'
b'190904-03:45:13,906 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_fig0".\n'
b'190904-03:45:13,907 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_fig1".\n'
b'190904-03:45:13,908 nipype.workflow INFO:\n'
b'\t [Node] Finished "_rename_matrix_node0".\n'
b'190904-03:45:13,909 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_fig2".\n'
b'190904-03:45:15,877 nipype.workflow INFO:\n'
b'\t [Job 8] Completed (_ds_correlation_fig0).\n'
b'190904-03:45:15,878 nipype.workflow INFO:\n'
b'\t [Job 9] Completed (_ds_correlation_fig1).\n'
b'190904-03:45:15,878 nipype.workflow INFO:\n'
b'\t [Job 10] Completed (_ds_correlation_fig2).\n'
b'190904-03:45:15,879 nipype.workflow INFO:\n'
b'\t [Job 11] Completed (_rename_matrix_node0).\n'
b'190904-03:45:15,880 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190904-03:45:15,903 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.ds_correlation_fig" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/ds_correlation_fig".\n'
b'190904-03:45:15,904 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_rename_matrix_node1" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/rename_matrix_node/mapflow/_rename_matrix_node1".\n'
b'190904-03:45:15,905 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_rename_matrix_node2" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/rename_matrix_node/mapflow/_rename_matrix_node2".\n'
b'190904-03:45:15,906 nipype.workflow INFO:\n'
b'\t [Node] Running "_rename_matrix_node1" ("nipype.interfaces.utility.wrappers.Function")\n'
b'190904-03:45:15,907 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_fig0" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/ds_correlation_fig/mapflow/_ds_correlation_fig0".\n'
b'190904-03:45:15,907 nipype.workflow INFO:\n'
b'\t [Node] Running "_rename_matrix_node2" ("nipype.interfaces.utility.wrappers.Function")\n'
b'190904-03:45:15,909 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_fig0" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190904-03:45:15,910 nipype.workflow INFO:\n'
b'\t [Node] Finished "_rename_matrix_node1".\n'
b'190904-03:45:15,911 nipype.workflow INFO:\n'
b'\t [Node] Finished "_rename_matrix_node2".\n'
b'190904-03:45:15,916 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_fig0".\n'
b'190904-03:45:15,916 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_fig1" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/ds_correlation_fig/mapflow/_ds_correlation_fig1".\n'
b'190904-03:45:15,919 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_fig1" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190904-03:45:15,925 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_fig1".\n'
b'190904-03:45:15,926 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_fig2" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/ds_correlation_fig/mapflow/_ds_correlation_fig2".\n'
b'190904-03:45:15,929 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_fig2" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190904-03:45:15,935 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_fig2".\n'
b'190904-03:45:15,938 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.ds_correlation_fig".\n'
b'190904-03:45:17,880 nipype.workflow INFO:\n'
b'\t [Job 2] Completed (nibetaseries_participant_wf.single_subject001_wf.ds_correlation_fig).\n'
b'190904-03:45:17,881 nipype.workflow INFO:\n'
b'\t [Job 12] Completed (_rename_matrix_node1).\n'
b'190904-03:45:17,881 nipype.workflow INFO:\n'
b'\t [Job 13] Completed (_rename_matrix_node2).\n'
b'190904-03:45:17,883 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190904-03:45:17,906 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.correlation_wf.rename_matrix_node" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/rename_matrix_node".\n'
b'190904-03:45:17,909 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_rename_matrix_node0" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/rename_matrix_node/mapflow/_rename_matrix_node0".\n'
b'190904-03:45:17,910 nipype.workflow INFO:\n'
b'\t [Node] Cached "_rename_matrix_node0" - collecting precomputed outputs\n'
b'190904-03:45:17,910 nipype.workflow INFO:\n'
b'\t [Node] "_rename_matrix_node0" found cached.\n'
b'190904-03:45:17,912 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_rename_matrix_node1" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/rename_matrix_node/mapflow/_rename_matrix_node1".\n'
b'190904-03:45:17,913 nipype.workflow INFO:\n'
b'\t [Node] Cached "_rename_matrix_node1" - collecting precomputed outputs\n'
b'190904-03:45:17,913 nipype.workflow INFO:\n'
b'\t [Node] "_rename_matrix_node1" found cached.\n'
b'190904-03:45:17,914 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_rename_matrix_node2" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/rename_matrix_node/mapflow/_rename_matrix_node2".\n'
b'190904-03:45:17,914 nipype.workflow INFO:\n'
b'\t [Node] Cached "_rename_matrix_node2" - collecting precomputed outputs\n'
b'190904-03:45:17,914 nipype.workflow INFO:\n'
b'\t [Node] "_rename_matrix_node2" found cached.\n'
b'190904-03:45:17,917 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.correlation_wf.rename_matrix_node".\n'
b'190904-03:45:19,882 nipype.workflow INFO:\n'
b'\t [Job 3] Completed (nibetaseries_participant_wf.single_subject001_wf.correlation_wf.rename_matrix_node).\n'
b'190904-03:45:19,884 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190904-03:45:21,885 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190904-03:45:21,906 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix0" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/ds_correlation_matrix/mapflow/_ds_correlation_matrix0".\n'
b'190904-03:45:21,908 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix1" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/ds_correlation_matrix/mapflow/_ds_correlation_matrix1".\n'
b'190904-03:45:21,908 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix0" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190904-03:45:21,909 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix2" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/ds_correlation_matrix/mapflow/_ds_correlation_matrix2".\n'
b'190904-03:45:21,909 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix1" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190904-03:45:21,911 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix2" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190904-03:45:21,912 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix0".\n'
b'190904-03:45:21,913 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix1".\n'
b'190904-03:45:21,914 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix2".\n'
b'190904-03:45:23,886 nipype.workflow INFO:\n'
b'\t [Job 14] Completed (_ds_correlation_matrix0).\n'
b'190904-03:45:23,887 nipype.workflow INFO:\n'
b'\t [Job 15] Completed (_ds_correlation_matrix1).\n'
b'190904-03:45:23,887 nipype.workflow INFO:\n'
b'\t [Job 16] Completed (_ds_correlation_matrix2).\n'
b'190904-03:45:23,889 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190904-03:45:23,916 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.ds_correlation_matrix" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/ds_correlation_matrix".\n'
b'190904-03:45:23,920 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix0" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/ds_correlation_matrix/mapflow/_ds_correlation_matrix0".\n'
b'190904-03:45:23,922 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix0" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190904-03:45:23,926 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix0".\n'
b'190904-03:45:23,927 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix1" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/ds_correlation_matrix/mapflow/_ds_correlation_matrix1".\n'
b'190904-03:45:23,929 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix1" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190904-03:45:23,933 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix1".\n'
b'190904-03:45:23,934 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix2" in "/tmp/tmpfobybnla/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/e008472dc43ca0472a118f349c7de768c0fda7bc/ds_correlation_matrix/mapflow/_ds_correlation_matrix2".\n'
b'190904-03:45:23,936 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix2" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190904-03:45:23,939 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix2".\n'
b'190904-03:45:23,941 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.ds_correlation_matrix".\n'
b'190904-03:45:25,888 nipype.workflow INFO:\n'
b'\t [Job 4] Completed (nibetaseries_participant_wf.single_subject001_wf.ds_correlation_matrix).\n'
b'190904-03:45:25,890 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 0 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.3.1/lib/python3.7/site-packages/nibetaseries/interfaces/nilearn.py:83: RuntimeWarning: invalid value encountered in greater\n'
b' n_lines = int(np.sum(connmat > 0) / 2)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.3.1/lib/python3.7/site-packages/nibetaseries/interfaces/nilearn.py:83: RuntimeWarning: invalid value encountered in greater\n'
b' n_lines = int(np.sum(connmat > 0) / 2)\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.3.1/lib/python3.7/site-packages/nibetaseries/interfaces/nilearn.py:83: RuntimeWarning: invalid value encountered in greater\n'
b' n_lines = int(np.sum(connmat > 0) / 2)\n'
Observe generated outputs¶
list_files(data_dir)
Out:
tmpfobybnla/
ds000164/
T1w.json
README
task-stroop_bold.json
dataset_description.json
task-stroop_events.json
CHANGES
derivatives/
NiBetaSeries/
nibetaseries/
sub-001/
func/
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc_trialtype-congruent_matrix.tsv
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc_trialtype-congruent_fig.svg
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc_trialtype-neutral_fig.svg
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc_trialtype-incongruent_fig.svg
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc_trialtype-incongruent_matrix.tsv
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc_trialtype-neutral_matrix.tsv
logs/
data/
Schaefer2018_100Parcels_7Networks_order.tsv
Schaefer2018_100Parcels_7Networks_order.txt
Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz
work/
NiBetaSeries_work/
nibetaseries_participant_wf/
graph1.json
d3.js
index.html
graph.json
single_subject001_wf/
e008472dc43ca0472a118f349c7de768c0fda7bc/
ds_correlation_matrix/
_0x4a94eec593110fc086ee9933dcbb5045.json
_node.pklz
result_ds_correlation_matrix.pklz
_inputs.pklz
_report/
report.rst
mapflow/
_ds_correlation_matrix0/
_node.pklz
result__ds_correlation_matrix0.pklz
_inputs.pklz
_0x96f5a85e42e889c658760c8651346bef.json
_report/
report.rst
_ds_correlation_matrix1/
_node.pklz
_inputs.pklz
_0x0d3c205a83cbe8bd1691788f56e2b8d6.json
result__ds_correlation_matrix1.pklz
_report/
report.rst
_ds_correlation_matrix2/
_node.pklz
_inputs.pklz
result__ds_correlation_matrix2.pklz
_0x4bf18f0bf571b272fd615d5777f654d9.json
_report/
report.rst
ds_correlation_fig/
_0x7f10b1ac985e7136c33d3fbf90535365.json
_node.pklz
_inputs.pklz
result_ds_correlation_fig.pklz
_report/
report.rst
mapflow/
_ds_correlation_fig0/
_node.pklz
_inputs.pklz
result__ds_correlation_fig0.pklz
_0xf7b486a0401c00759439067797130ac8.json
_report/
report.rst
_ds_correlation_fig2/
_node.pklz
_inputs.pklz
result__ds_correlation_fig2.pklz
_0x914d84eea107c2cad4ed5560be74255d.json
_report/
report.rst
_ds_correlation_fig1/
_node.pklz
_inputs.pklz
result__ds_correlation_fig1.pklz
_0x3528265cebeff77e7ef7a9a1d953f88a.json
_report/
report.rst
correlation_wf/
e008472dc43ca0472a118f349c7de768c0fda7bc/
atlas_corr_node/
_node.pklz
result_atlas_corr_node.pklz
_inputs.pklz
_0xbc5c884c38625f4e661d6e10444614e0.json
_report/
report.rst
mapflow/
_atlas_corr_node2/
fisher_z_correlation.tsv
_node.pklz
_inputs.pklz
result__atlas_corr_node2.pklz
incongruent.svg
_0x9a19a6a43af7abd35a3231ff77e1a527.json
_report/
report.rst
_atlas_corr_node0/
fisher_z_correlation.tsv
_node.pklz
_inputs.pklz
_0x8a70d8c38afee452b9fee82235dd0b32.json
neutral.svg
result__atlas_corr_node0.pklz
_report/
report.rst
_atlas_corr_node1/
fisher_z_correlation.tsv
_node.pklz
_inputs.pklz
_0x8583197fd41212b8d8194c97f0286110.json
result__atlas_corr_node1.pklz
congruent.svg
_report/
report.rst
rename_matrix_node/
_node.pklz
result_rename_matrix_node.pklz
_inputs.pklz
_0xb953ddf48447d6948f0cbbbf6f27d42c.json
_report/
report.rst
mapflow/
_rename_matrix_node1/
_node.pklz
_inputs.pklz
result__rename_matrix_node1.pklz
correlation-matrix_trialtype-congruent.tsv
_0xbc05af300d397c7aaf1c3978f81b1956.json
_report/
report.rst
_rename_matrix_node2/
_0xbd2638de770e93c8a78c51b14f45e93c.json
_node.pklz
_inputs.pklz
correlation-matrix_trialtype-incongruent.tsv
result__rename_matrix_node2.pklz
_report/
report.rst
_rename_matrix_node0/
_0x4fe7f4c207fb404faa4b3a4e606c7951.json
_node.pklz
_inputs.pklz
result__rename_matrix_node0.pklz
correlation-matrix_trialtype-neutral.tsv
_report/
report.rst
betaseries_wf/
e008472dc43ca0472a118f349c7de768c0fda7bc/
betaseries_node/
_node.pklz
_inputs.pklz
betaseries_trialtype-neutral.nii.gz
betaseries_trialtype-congruent.nii.gz
_0x12cd3ac1a94d484f3efb345680c64c01.json
betaseries_trialtype-incongruent.nii.gz
result_betaseries_node.pklz
_report/
report.rst
fmriprep/
sub-001/
func/
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_brainmask.nii.gz
sub-001_task-stroop_bold_confounds.tsv
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz
sub-001/
anat/
sub-001_T1w.nii.gz
func/
sub-001_task-stroop_bold.nii.gz
sub-001_task-stroop_events.tsv
Collect results¶
corr_mat_path = os.path.join(out_dir, "NiBetaSeries", "nibetaseries", "sub-001", "func")
trial_types = ['congruent', 'incongruent', 'neutral']
filename_template = "sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc_trialtype-{trial_type}_matrix.tsv"
pd_dict = {}
for trial_type in trial_types:
file_path = os.path.join(corr_mat_path, filename_template.format(trial_type=trial_type))
pd_dict[trial_type] = pd.read_csv(file_path, sep='\t', na_values="n/a", index_col=0)
# display example matrix
print(pd_dict[trial_type].head())
Out:
LH_Vis_1 LH_Vis_2 ... RH_Default_PCC_1 RH_Default_PCC_2
LH_Vis_1 NaN 0.092135 ... 0.095624 0.016799
LH_Vis_2 0.092135 NaN ... -0.119613 -0.007679
LH_Vis_3 -0.003990 0.216346 ... 0.202673 0.177828
LH_Vis_4 0.075498 -0.088788 ... -0.019256 -0.034034
LH_Vis_5 0.314494 0.354525 ... -0.235334 0.032317
[5 rows x 100 columns]
Graph the results¶
fig, axes = plt.subplots(nrows=3, ncols=1, sharex=True, sharey=True, figsize=(10, 30),
gridspec_kw={'wspace': 0.025, 'hspace': 0.075})
cbar_ax = fig.add_axes([.91, .3, .03, .4])
r = 0
for trial_type, df in pd_dict.items():
g = sns.heatmap(df, ax=axes[r], vmin=-.5, vmax=1., square=True,
cbar=True, cbar_ax=cbar_ax)
axes[r].set_title(trial_type)
# iterate over rows
r += 1
plt.tight_layout()
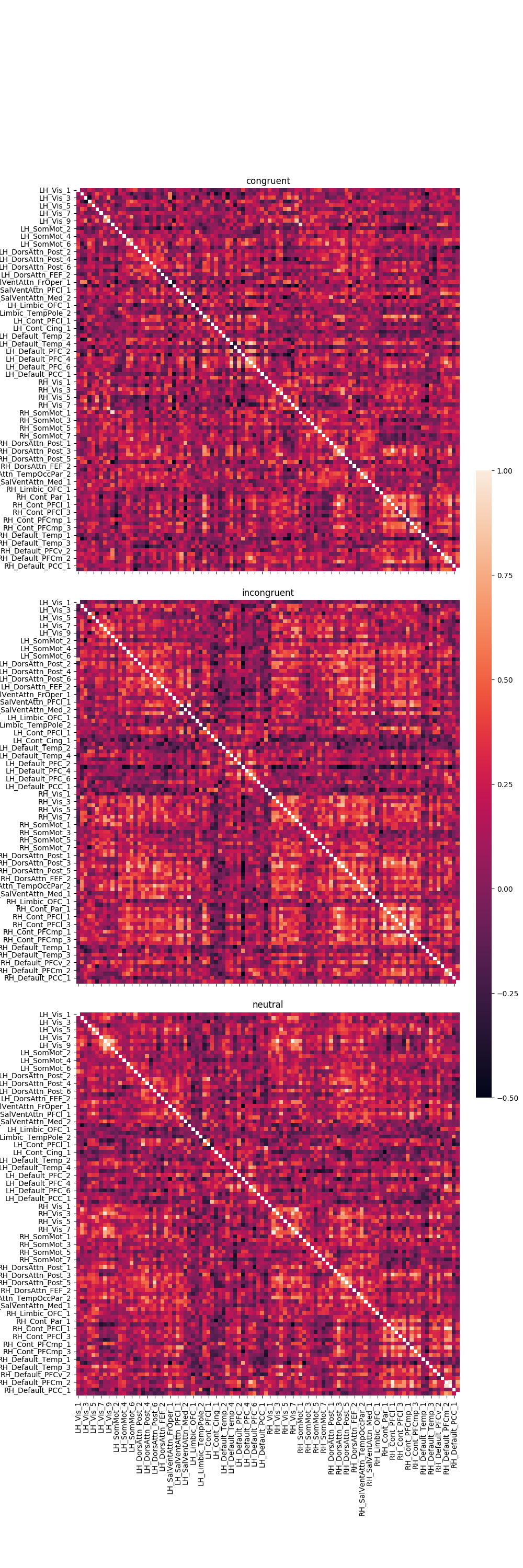
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.3.1/lib/python3.7/site-packages/matplotlib/figure.py:2299: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
warnings.warn("This figure includes Axes that are not compatible "
References¶
- notebook-1
Timothy D Verstynen. The organization and dynamics of corticostriatal pathways link the medial orbitofrontal cortex to future behavioral responses. Journal of Neurophysiology, 112(10):2457–2469, 2014. URL: https://doi.org/10.1152/jn.00221.2014, doi:10.1152/jn.00221.2014.
Total running time of the script: ( 1 minutes 54.277 seconds)