Note
Click here to download the full example code or to run this example in your browser via Binder
Running NiBetaSeries¶
This example runs through a basic call of NiBetaSeries using
the commandline entry point nibs.
While this example is using python, typically nibs will be
called directly on the commandline.
Import all the necessary packages¶
import tempfile # make a temporary directory for files
import os # interact with the filesystem
import urllib.request # grad data from internet
import tarfile # extract files from tar
from subprocess import Popen, PIPE, STDOUT # enable calling commandline
import matplotlib.pyplot as plt # manipulate figures
import seaborn as sns # display results
import pandas as pd # manipulate tabular data
import nibabel as nib # load the beta maps in python
from nilearn import plotting # plot nifti images
Download relevant data from ds000164 (and Atlas Files)¶
The subject data came from openneuro [notebook-1]. The atlas data came from a recently published parcellation in a publically accessible github repository.
# atlas github repo for reference:
"""https://github.com/ThomasYeoLab/CBIG/raw/master/stable_projects/\
brain_parcellation/Schaefer2018_LocalGlobal/Parcellations/MNI/"""
data_dir = tempfile.mkdtemp()
print('Our working directory: {}'.format(data_dir))
# download the tar data
url = "https://www.dropbox.com/s/fvtyld08srwl3x9/ds000164-test_v2.tar.gz?dl=1"
tar_file = os.path.join(data_dir, "ds000164.tar.gz")
u = urllib.request.urlopen(url)
data = u.read()
u.close()
# write tar data to file
with open(tar_file, "wb") as f:
f.write(data)
# extract the data
tar = tarfile.open(tar_file, mode='r|gz')
tar.extractall(path=data_dir)
os.remove(tar_file)
Out:
Our working directory: /tmp/tmpsh4asxeu
Display the minimal dataset necessary to run nibs¶
# https://stackoverflow.com/questions/9727673/list-directory-tree-structure-in-python
def list_files(startpath):
for root, dirs, files in os.walk(startpath):
level = root.replace(startpath, '').count(os.sep)
indent = ' ' * 4 * (level)
print('{}{}/'.format(indent, os.path.basename(root)))
subindent = ' ' * 4 * (level + 1)
for f in files:
print('{}{}'.format(subindent, f))
list_files(data_dir)
Out:
tmpsh4asxeu/
ds000164/
CHANGES
dataset_description.json
T1w.json
README
task-stroop_bold.json
task-stroop_events.json
derivatives/
fmriprep/
dataset_description.json
sub-001/
func/
sub-001_task-stroop_desc-confounds_regressors.tsv
sub-001_task-stroop_space-MNI152NLin2009cAsym_desc-brain_mask.nii.gz
sub-001_task-stroop_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
data/
Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz
Schaefer2018_100Parcels_7Networks_order.txt
sub-001/
anat/
sub-001_T1w.nii.gz
func/
sub-001_task-stroop_bold.nii.gz
sub-001_task-stroop_events.tsv
Manipulate events file so it satifies assumptions¶
1. the correct column has 1’s and 0’s corresponding to correct and incorrect, respectively. 2. the condition column is renamed to trial_type nibs currently depends on the “correct” column being binary and the “trial_type” column to contain the trial types of interest.
read the file¶
events_file = os.path.join(data_dir,
"ds000164",
"sub-001",
"func",
"sub-001_task-stroop_events.tsv")
events_df = pd.read_csv(events_file, sep='\t', na_values="n/a")
print(events_df.head())
Out:
onset duration correct condition response_time
0 0.342 1 Y neutral 1.186
1 3.345 1 Y congruent 0.667
2 12.346 1 Y congruent 0.614
3 15.349 1 Y neutral 0.696
4 18.350 1 Y neutral 0.752
replace condition with trial_type¶
events_df.rename({"condition": "trial_type"}, axis='columns', inplace=True)
print(events_df.head())
Out:
onset duration correct trial_type response_time
0 0.342 1 Y neutral 1.186
1 3.345 1 Y congruent 0.667
2 12.346 1 Y congruent 0.614
3 15.349 1 Y neutral 0.696
4 18.350 1 Y neutral 0.752
save the file¶
events_df.to_csv(events_file, sep="\t", na_rep="n/a", index=False)
Manipulate the region order file¶
There are several adjustments to the atlas file that need to be completed before we can pass it into nibs. Importantly, the relevant column names MUST be named “index” and “regions”. “index” refers to which integer within the file corresponds to which region in the atlas nifti file. “regions” refers the name of each region in the atlas nifti file.
read the atlas file¶
atlas_txt = os.path.join(data_dir,
"ds000164",
"derivatives",
"data",
"Schaefer2018_100Parcels_7Networks_order.txt")
atlas_df = pd.read_csv(atlas_txt, sep="\t", header=None)
print(atlas_df.head())
Out:
0 1 2 3 4 5
0 1 7Networks_LH_Vis_1 120 18 131 0
1 2 7Networks_LH_Vis_2 120 18 132 0
2 3 7Networks_LH_Vis_3 120 18 133 0
3 4 7Networks_LH_Vis_4 120 18 135 0
4 5 7Networks_LH_Vis_5 120 18 136 0
drop color coding columns¶
atlas_df.drop([2, 3, 4, 5], axis='columns', inplace=True)
print(atlas_df.head())
Out:
0 1
0 1 7Networks_LH_Vis_1
1 2 7Networks_LH_Vis_2
2 3 7Networks_LH_Vis_3
3 4 7Networks_LH_Vis_4
4 5 7Networks_LH_Vis_5
rename columns with the approved headings: “index” and “regions”¶
atlas_df.rename({0: 'index', 1: 'regions'}, axis='columns', inplace=True)
print(atlas_df.head())
Out:
index regions
0 1 7Networks_LH_Vis_1
1 2 7Networks_LH_Vis_2
2 3 7Networks_LH_Vis_3
3 4 7Networks_LH_Vis_4
4 5 7Networks_LH_Vis_5
remove prefix “7Networks”¶
atlas_df.replace(regex={'7Networks_(.*)': '\\1'}, inplace=True)
print(atlas_df.head())
Out:
index regions
0 1 LH_Vis_1
1 2 LH_Vis_2
2 3 LH_Vis_3
3 4 LH_Vis_4
4 5 LH_Vis_5
write out the file as .tsv¶
atlas_tsv = atlas_txt.replace(".txt", ".tsv")
atlas_df.to_csv(atlas_tsv, sep="\t", index=False)
Run nibs¶
This demonstration mimics how you would use nibs on the command line If you only wanted the beta maps and not the correlation matrices, do not include the atlas (-a) and lookup table options (-l)
out_dir = os.path.join(data_dir, "ds000164", "derivatives")
work_dir = os.path.join(out_dir, "work")
atlas_mni_file = os.path.join(data_dir,
"ds000164",
"derivatives",
"data",
"Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz")
cmd = """\
nibs -c WhiteMatter CSF \
--participant-label 001 \
--estimator lsa \
--hrf-model glover \
-w {work_dir} \
-a {atlas_mni_file} \
-l {atlas_tsv} \
{bids_dir} \
fmriprep \
{out_dir} \
participant
""".format(atlas_mni_file=atlas_mni_file,
atlas_tsv=atlas_tsv,
bids_dir=os.path.join(data_dir, "ds000164"),
out_dir=out_dir,
work_dir=work_dir)
# Since we cannot run bash commands inside this tutorial
# we are printing the actual bash command so you can see it
# in the output
print("The Example Command:\n", cmd)
# call nibs
p = Popen(cmd, shell=True, stdout=PIPE, stderr=STDOUT)
while True:
line = p.stdout.readline()
if not line:
break
print(line)
Out:
The Example Command:
nibs -c WhiteMatter CSF --participant-label 001 --estimator lsa --hrf-model glover -w /tmp/tmpsh4asxeu/ds000164/derivatives/work -a /tmp/tmpsh4asxeu/ds000164/derivatives/data/Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz -l /tmp/tmpsh4asxeu/ds000164/derivatives/data/Schaefer2018_100Parcels_7Networks_order.tsv /tmp/tmpsh4asxeu/ds000164 fmriprep /tmp/tmpsh4asxeu/ds000164/derivatives participant
b'200409-23:23:55,53 nipype.utils INFO:\n'
b'\t No new version available.\n'
b'200409-23:23:55,117 nipype.workflow INFO:\n'
b"\t Workflow nibetaseries_participant_wf settings: ['check', 'execution', 'logging', 'monitoring']\n"
b'200409-23:23:55,131 nipype.workflow INFO:\n'
b'\t Running in parallel.\n'
b'200409-23:23:55,133 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b"/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.6.0/lib/python3.7/site-packages/bids/layout/models.py:157: UserWarning: Accessing entities as attributes is deprecated as of 0.7. Please use the .entities dictionary instead (i.e., .entities['task'] instead of .task.\n"
b' % (attr, attr))\n'
b"/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.6.0/lib/python3.7/site-packages/bids/layout/models.py:157: UserWarning: Accessing entities as attributes is deprecated as of 0.7. Please use the .entities dictionary instead (i.e., .entities['space'] instead of .space.\n"
b' % (attr, attr))\n'
b'200409-23:23:55,187 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.betaseries_wf.betaseries_node" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/betaseries_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/betaseries_node".\n'
b'200409-23:23:55,192 nipype.workflow INFO:\n'
b'\t [Node] Running "betaseries_node" ("nibetaseries.interfaces.nistats.LSABetaSeries")\n'
b'200409-23:23:57,136 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 1 tasks, and 0 jobs ready. Free memory (GB): 2.80/3.00, Free processors: 0/1.\n'
b' Currently running:\n'
b' * nibetaseries_participant_wf.single_subject001_wf.betaseries_wf.betaseries_node\n'
b'200409-23:23:59,489 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.betaseries_wf.betaseries_node".\n'
b'200409-23:24:01,139 nipype.workflow INFO:\n'
b'\t [Job 0] Completed (nibetaseries_participant_wf.single_subject001_wf.betaseries_wf.betaseries_node).\n'
b'200409-23:24:01,142 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 2 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:03,142 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 6 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:03,193 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_betaseries_file0" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_betaseries_file/mapflow/_ds_betaseries_file0".\n'
b'200409-23:24:03,195 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_betaseries_file0" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:24:03,205 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_betaseries_file0".\n'
b'200409-23:24:05,143 nipype.workflow INFO:\n'
b'\t [Job 7] Completed (_ds_betaseries_file0).\n'
b'200409-23:24:05,145 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 5 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:05,192 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_betaseries_file1" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_betaseries_file/mapflow/_ds_betaseries_file1".\n'
b'200409-23:24:05,193 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_betaseries_file1" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:24:05,201 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_betaseries_file1".\n'
b'200409-23:24:07,145 nipype.workflow INFO:\n'
b'\t [Job 8] Completed (_ds_betaseries_file1).\n'
b'200409-23:24:07,147 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 4 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:07,192 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_betaseries_file2" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_betaseries_file/mapflow/_ds_betaseries_file2".\n'
b'200409-23:24:07,193 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_betaseries_file2" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:24:07,201 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_betaseries_file2".\n'
b'200409-23:24:09,147 nipype.workflow INFO:\n'
b'\t [Job 9] Completed (_ds_betaseries_file2).\n'
b'200409-23:24:09,150 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 4 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:09,195 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.ds_betaseries_file" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_betaseries_file".\n'
b'200409-23:24:09,198 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_betaseries_file0" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_betaseries_file/mapflow/_ds_betaseries_file0".\n'
b'200409-23:24:09,200 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_betaseries_file0" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:24:09,208 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_betaseries_file0".\n'
b'200409-23:24:09,209 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_betaseries_file1" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_betaseries_file/mapflow/_ds_betaseries_file1".\n'
b'200409-23:24:09,211 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_betaseries_file1" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:24:09,218 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_betaseries_file1".\n'
b'200409-23:24:09,219 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_betaseries_file2" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_betaseries_file/mapflow/_ds_betaseries_file2".\n'
b'200409-23:24:09,221 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_betaseries_file2" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:24:09,229 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_betaseries_file2".\n'
b'200409-23:24:09,230 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.ds_betaseries_file".\n'
b'200409-23:24:11,150 nipype.workflow INFO:\n'
b'\t [Job 1] Completed (nibetaseries_participant_wf.single_subject001_wf.ds_betaseries_file).\n'
b'200409-23:24:11,152 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:11,196 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_censor_volumes0" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/censor_volumes/mapflow/_censor_volumes0".\n'
b'200409-23:24:11,197 nipype.workflow INFO:\n'
b'\t [Node] Running "_censor_volumes0" ("nibetaseries.interfaces.nilearn.CensorVolumes")\n'
b'200409-23:24:11,258 nipype.workflow INFO:\n'
b'\t [Node] Finished "_censor_volumes0".\n'
b'200409-23:24:13,152 nipype.workflow INFO:\n'
b'\t [Job 10] Completed (_censor_volumes0).\n'
b'200409-23:24:13,154 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 2 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:13,198 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_censor_volumes1" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/censor_volumes/mapflow/_censor_volumes1".\n'
b'200409-23:24:13,200 nipype.workflow INFO:\n'
b'\t [Node] Running "_censor_volumes1" ("nibetaseries.interfaces.nilearn.CensorVolumes")\n'
b'200409-23:24:13,263 nipype.workflow INFO:\n'
b'\t [Node] Finished "_censor_volumes1".\n'
b'200409-23:24:15,154 nipype.workflow INFO:\n'
b'\t [Job 11] Completed (_censor_volumes1).\n'
b'200409-23:24:15,156 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:15,203 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_censor_volumes2" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/censor_volumes/mapflow/_censor_volumes2".\n'
b'200409-23:24:15,205 nipype.workflow INFO:\n'
b'\t [Node] Running "_censor_volumes2" ("nibetaseries.interfaces.nilearn.CensorVolumes")\n'
b'200409-23:24:15,257 nipype.workflow INFO:\n'
b'\t [Node] Finished "_censor_volumes2".\n'
b'200409-23:24:17,155 nipype.workflow INFO:\n'
b'\t [Job 12] Completed (_censor_volumes2).\n'
b'200409-23:24:17,157 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:17,209 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.censor_volumes" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/censor_volumes".\n'
b'200409-23:24:17,213 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_censor_volumes0" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/censor_volumes/mapflow/_censor_volumes0".\n'
b'200409-23:24:17,214 nipype.workflow INFO:\n'
b'\t [Node] Cached "_censor_volumes0" - collecting precomputed outputs\n'
b'200409-23:24:17,215 nipype.workflow INFO:\n'
b'\t [Node] "_censor_volumes0" found cached.\n'
b'200409-23:24:17,216 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_censor_volumes1" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/censor_volumes/mapflow/_censor_volumes1".\n'
b'200409-23:24:17,217 nipype.workflow INFO:\n'
b'\t [Node] Cached "_censor_volumes1" - collecting precomputed outputs\n'
b'200409-23:24:17,217 nipype.workflow INFO:\n'
b'\t [Node] "_censor_volumes1" found cached.\n'
b'200409-23:24:17,218 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_censor_volumes2" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/censor_volumes/mapflow/_censor_volumes2".\n'
b'200409-23:24:17,219 nipype.workflow INFO:\n'
b'\t [Node] Cached "_censor_volumes2" - collecting precomputed outputs\n'
b'200409-23:24:17,219 nipype.workflow INFO:\n'
b'\t [Node] "_censor_volumes2" found cached.\n'
b'200409-23:24:17,220 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.censor_volumes".\n'
b'200409-23:24:19,157 nipype.workflow INFO:\n'
b'\t [Job 2] Completed (nibetaseries_participant_wf.single_subject001_wf.censor_volumes).\n'
b'200409-23:24:19,160 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:19,210 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.check_beta_series_list" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/check_beta_series_list".\n'
b'200409-23:24:19,214 nipype.workflow INFO:\n'
b'\t [Node] Running "check_beta_series_list" ("nipype.interfaces.utility.wrappers.Function")\n'
b'200409-23:24:19,220 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.check_beta_series_list".\n'
b'200409-23:24:21,160 nipype.workflow INFO:\n'
b'\t [Job 3] Completed (nibetaseries_participant_wf.single_subject001_wf.check_beta_series_list).\n'
b'200409-23:24:21,163 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:23,164 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:23,211 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node0" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/atlas_corr_node/mapflow/_atlas_corr_node0".\n'
b'200409-23:24:23,213 nipype.workflow INFO:\n'
b'\t [Node] Running "_atlas_corr_node0" ("nibetaseries.interfaces.nilearn.AtlasConnectivity")\n'
b'200409-23:24:25,164 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 1 tasks, and 2 jobs ready. Free memory (GB): 2.80/3.00, Free processors: 0/1.\n'
b' Currently running:\n'
b' * _atlas_corr_node0\n'
b'[NiftiLabelsMasker.fit_transform] loading data from /tmp/tmpsh4asxeu/ds000164/derivatives/data/Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz\n'
b'Resampling labels\n'
b'[NiftiLabelsMasker.transform_single_imgs] Loading data from /tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/betaseries_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/betaseries_node/desc-neutral_betase\n'
b'[NiftiLabelsMasker.transform_single_imgs] Extracting region signals\n'
b'[NiftiLabelsMasker.transform_single_imgs] Cleaning extracted signals\n'
b'200409-23:24:29,343 nipype.workflow INFO:\n'
b'\t [Node] Finished "_atlas_corr_node0".\n'
b'200409-23:24:31,169 nipype.workflow INFO:\n'
b'\t [Job 13] Completed (_atlas_corr_node0).\n'
b'200409-23:24:31,172 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 2 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:31,214 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node1" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/atlas_corr_node/mapflow/_atlas_corr_node1".\n'
b'200409-23:24:31,216 nipype.workflow INFO:\n'
b'\t [Node] Running "_atlas_corr_node1" ("nibetaseries.interfaces.nilearn.AtlasConnectivity")\n'
b'200409-23:24:33,171 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 1 tasks, and 1 jobs ready. Free memory (GB): 2.80/3.00, Free processors: 0/1.\n'
b' Currently running:\n'
b' * _atlas_corr_node1\n'
b'[NiftiLabelsMasker.fit_transform] loading data from /tmp/tmpsh4asxeu/ds000164/derivatives/data/Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz\n'
b'Resampling labels\n'
b'[NiftiLabelsMasker.transform_single_imgs] Loading data from /tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/betaseries_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/betaseries_node/desc-congruent_beta\n'
b'[NiftiLabelsMasker.transform_single_imgs] Extracting region signals\n'
b'[NiftiLabelsMasker.transform_single_imgs] Cleaning extracted signals\n'
b'200409-23:24:37,701 nipype.workflow INFO:\n'
b'\t [Node] Finished "_atlas_corr_node1".\n'
b'200409-23:24:39,177 nipype.workflow INFO:\n'
b'\t [Job 14] Completed (_atlas_corr_node1).\n'
b'200409-23:24:39,179 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:39,226 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node2" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/atlas_corr_node/mapflow/_atlas_corr_node2".\n'
b'200409-23:24:39,228 nipype.workflow INFO:\n'
b'\t [Node] Running "_atlas_corr_node2" ("nibetaseries.interfaces.nilearn.AtlasConnectivity")\n'
b'200409-23:24:41,179 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 1 tasks, and 0 jobs ready. Free memory (GB): 2.80/3.00, Free processors: 0/1.\n'
b' Currently running:\n'
b' * _atlas_corr_node2\n'
b'[NiftiLabelsMasker.fit_transform] loading data from /tmp/tmpsh4asxeu/ds000164/derivatives/data/Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz\n'
b'Resampling labels\n'
b'[NiftiLabelsMasker.transform_single_imgs] Loading data from /tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/betaseries_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/betaseries_node/desc-incongruent_be\n'
b'[NiftiLabelsMasker.transform_single_imgs] Extracting region signals\n'
b'[NiftiLabelsMasker.transform_single_imgs] Cleaning extracted signals\n'
b'200409-23:24:45,416 nipype.workflow INFO:\n'
b'\t [Node] Finished "_atlas_corr_node2".\n'
b'200409-23:24:47,184 nipype.workflow INFO:\n'
b'\t [Job 15] Completed (_atlas_corr_node2).\n'
b'200409-23:24:47,186 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:47,232 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.correlation_wf.atlas_corr_node" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/atlas_corr_node".\n'
b'200409-23:24:47,236 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node0" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/atlas_corr_node/mapflow/_atlas_corr_node0".\n'
b'200409-23:24:47,237 nipype.workflow INFO:\n'
b'\t [Node] Cached "_atlas_corr_node0" - collecting precomputed outputs\n'
b'200409-23:24:47,237 nipype.workflow INFO:\n'
b'\t [Node] "_atlas_corr_node0" found cached.\n'
b'200409-23:24:47,238 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node1" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/atlas_corr_node/mapflow/_atlas_corr_node1".\n'
b'200409-23:24:47,239 nipype.workflow INFO:\n'
b'\t [Node] Cached "_atlas_corr_node1" - collecting precomputed outputs\n'
b'200409-23:24:47,240 nipype.workflow INFO:\n'
b'\t [Node] "_atlas_corr_node1" found cached.\n'
b'200409-23:24:47,241 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node2" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/atlas_corr_node/mapflow/_atlas_corr_node2".\n'
b'200409-23:24:47,242 nipype.workflow INFO:\n'
b'\t [Node] Cached "_atlas_corr_node2" - collecting precomputed outputs\n'
b'200409-23:24:47,242 nipype.workflow INFO:\n'
b'\t [Node] "_atlas_corr_node2" found cached.\n'
b'200409-23:24:47,243 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.correlation_wf.atlas_corr_node".\n'
b'200409-23:24:49,187 nipype.workflow INFO:\n'
b'\t [Job 4] Completed (nibetaseries_participant_wf.single_subject001_wf.correlation_wf.atlas_corr_node).\n'
b'200409-23:24:49,190 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 2 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:51,189 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 6 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:51,236 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_fig0" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_correlation_fig/mapflow/_ds_correlation_fig0".\n'
b'200409-23:24:51,240 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_fig0" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:24:51,249 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_fig0".\n'
b'200409-23:24:53,191 nipype.workflow INFO:\n'
b'\t [Job 16] Completed (_ds_correlation_fig0).\n'
b'200409-23:24:53,194 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 5 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:53,239 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_fig1" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_correlation_fig/mapflow/_ds_correlation_fig1".\n'
b'200409-23:24:53,241 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_fig1" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:24:53,250 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_fig1".\n'
b'200409-23:24:55,193 nipype.workflow INFO:\n'
b'\t [Job 17] Completed (_ds_correlation_fig1).\n'
b'200409-23:24:55,195 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 4 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:55,244 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_fig2" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_correlation_fig/mapflow/_ds_correlation_fig2".\n'
b'200409-23:24:55,246 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_fig2" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:24:55,256 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_fig2".\n'
b'200409-23:24:57,196 nipype.workflow INFO:\n'
b'\t [Job 18] Completed (_ds_correlation_fig2).\n'
b'200409-23:24:57,197 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 4 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:57,242 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.ds_correlation_fig" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_correlation_fig".\n'
b'200409-23:24:57,246 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_fig0" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_correlation_fig/mapflow/_ds_correlation_fig0".\n'
b'200409-23:24:57,248 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_fig0" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:24:57,257 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_fig0".\n'
b'200409-23:24:57,258 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_fig1" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_correlation_fig/mapflow/_ds_correlation_fig1".\n'
b'200409-23:24:57,261 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_fig1" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:24:57,270 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_fig1".\n'
b'200409-23:24:57,273 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_fig2" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_correlation_fig/mapflow/_ds_correlation_fig2".\n'
b'200409-23:24:57,275 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_fig2" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:24:57,347 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_fig2".\n'
b'200409-23:24:57,350 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.ds_correlation_fig".\n'
b'200409-23:24:59,198 nipype.workflow INFO:\n'
b'\t [Job 5] Completed (nibetaseries_participant_wf.single_subject001_wf.ds_correlation_fig).\n'
b'200409-23:24:59,200 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:24:59,247 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix0" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_correlation_matrix/mapflow/_ds_correlation_matrix0".\n'
b'200409-23:24:59,249 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix0" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:24:59,256 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix0".\n'
b'200409-23:25:01,200 nipype.workflow INFO:\n'
b'\t [Job 19] Completed (_ds_correlation_matrix0).\n'
b'200409-23:25:01,202 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 2 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:25:01,247 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix1" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_correlation_matrix/mapflow/_ds_correlation_matrix1".\n'
b'200409-23:25:01,249 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix1" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:25:01,257 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix1".\n'
b'200409-23:25:03,202 nipype.workflow INFO:\n'
b'\t [Job 20] Completed (_ds_correlation_matrix1).\n'
b'200409-23:25:03,205 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:25:03,253 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix2" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_correlation_matrix/mapflow/_ds_correlation_matrix2".\n'
b'200409-23:25:03,254 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix2" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:25:03,262 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix2".\n'
b'200409-23:25:05,204 nipype.workflow INFO:\n'
b'\t [Job 21] Completed (_ds_correlation_matrix2).\n'
b'200409-23:25:05,207 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'200409-23:25:05,252 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.ds_correlation_matrix" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_correlation_matrix".\n'
b'200409-23:25:05,256 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix0" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_correlation_matrix/mapflow/_ds_correlation_matrix0".\n'
b'200409-23:25:05,258 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix0" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:25:05,265 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix0".\n'
b'200409-23:25:05,266 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix1" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_correlation_matrix/mapflow/_ds_correlation_matrix1".\n'
b'200409-23:25:05,268 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix1" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:25:05,275 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix1".\n'
b'200409-23:25:05,276 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix2" in "/tmp/tmpsh4asxeu/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/05f89e1eb2a40b75543a0ece936c065ae586c757/ds_correlation_matrix/mapflow/_ds_correlation_matrix2".\n'
b'200409-23:25:05,278 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix2" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'200409-23:25:05,285 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix2".\n'
b'200409-23:25:05,286 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.ds_correlation_matrix".\n'
b'200409-23:25:07,206 nipype.workflow INFO:\n'
b'\t [Job 6] Completed (nibetaseries_participant_wf.single_subject001_wf.ds_correlation_matrix).\n'
b'200409-23:25:07,209 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 0 jobs ready. Free memory (GB): 3.00/3.00, Free processors: 1/1.\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.6.0/lib/python3.7/site-packages/nibetaseries/interfaces/nistats.py:216: DeprecationWarning: The parameter "mask" will be removed in next release of Nistats. Please use the parameter "mask_img" instead.\n'
b' minimize_memory=False,\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 2 seconds\n'
b'\n'
b"/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.6.0/lib/python3.7/site-packages/nistats/regression.py:339: FutureWarning: 'resid' from RegressionResults has been deprecated and will be removed. Please use 'residuals' instead.\n"
b' FutureWarning,\n'
b"/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.6.0/lib/python3.7/site-packages/sklearn/feature_extraction/text.py:17: DeprecationWarning: Using or importing the ABCs from 'collections' instead of from 'collections.abc' is deprecated, and in 3.8 it will stop working\n"
b' from collections import Mapping, defaultdict\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.6.0/lib/python3.7/site-packages/nibetaseries/interfaces/nilearn.py:130: RuntimeWarning: invalid value encountered in greater\n'
b' n_lines = int(np.sum(connmat > 0) / 2)\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.6.0/lib/python3.7/site-packages/nibetaseries/interfaces/nilearn.py:130: RuntimeWarning: invalid value encountered in greater\n'
b' n_lines = int(np.sum(connmat > 0) / 2)\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.6.0/lib/python3.7/site-packages/nibetaseries/interfaces/nilearn.py:130: RuntimeWarning: invalid value encountered in greater\n'
b' n_lines = int(np.sum(connmat > 0) / 2)\n'
b'pandoc: Error running filter pandoc-citeproc:\n'
b"Could not find executable 'pandoc-citeproc'.\n"
b'Could not generate CITATION.html file:\n'
b'pandoc -s --bibliography /home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.6.0/lib/python3.7/site-packages/nibetaseries/data/references.bib --filter pandoc-citeproc --metadata pagetitle="NiBetaSeries citation boilerplate" /tmp/tmpsh4asxeu/ds000164/derivatives/nibetaseries/logs/CITATION.md -o /tmp/tmpsh4asxeu/ds000164/derivatives/nibetaseries/logs/CITATION.html\n'
Observe generated outputs¶
list_files(data_dir)
Out:
tmpsh4asxeu/
ds000164/
CHANGES
dataset_description.json
T1w.json
README
task-stroop_bold.json
task-stroop_events.json
derivatives/
work/
dbcache
NiBetaSeries_work/
nibetaseries_participant_wf/
graph1.json
graph.json
d3.js
index.html
single_subject001_wf/
correlation_wf/
05f89e1eb2a40b75543a0ece936c065ae586c757/
atlas_corr_node/
_inputs.pklz
result_atlas_corr_node.pklz
_0xb689bb6129c8848651e9f936ac5233c1.json
_node.pklz
_report/
report.rst
mapflow/
_atlas_corr_node0/
_inputs.pklz
result__atlas_corr_node0.pklz
_0xaba74adc5e8a4d23059b43d53e8628a0.json
desc-neutral_correlation.svg
_node.pklz
desc-neutral_correlation.tsv
_report/
report.rst
_atlas_corr_node1/
_inputs.pklz
desc-congruent_correlation.tsv
result__atlas_corr_node1.pklz
desc-congruent_correlation.svg
_node.pklz
_0x9fa7b666030de664cfc8c5fd40f3d74d.json
_report/
report.rst
_atlas_corr_node2/
_inputs.pklz
_0xeda7d619211a926224d07521d3995c54.json
_node.pklz
desc-incongruent_correlation.tsv
result__atlas_corr_node2.pklz
desc-incongruent_correlation.svg
_report/
report.rst
05f89e1eb2a40b75543a0ece936c065ae586c757/
censor_volumes/
_inputs.pklz
result_censor_volumes.pklz
_node.pklz
_0x9d3cc53ac9deb07b60d93d0e3d5e6a5c.json
_report/
report.rst
mapflow/
_censor_volumes1/
_inputs.pklz
_0x505f021ec6d1f6150005e304643a6a61.json
result__censor_volumes1.pklz
_node.pklz
_report/
report.rst
_censor_volumes0/
_inputs.pklz
_0xde9858f5b550513f65686dedb7335749.json
result__censor_volumes0.pklz
_node.pklz
_report/
report.rst
_censor_volumes2/
_0x78d9a5cce957a6e5e4976d7f34c7660b.json
_inputs.pklz
result__censor_volumes2.pklz
_node.pklz
_report/
report.rst
check_beta_series_list/
_inputs.pklz
result_check_beta_series_list.pklz
_node.pklz
_0x6adef7d6a32299c1a2356f35cfbdc6f2.json
_report/
report.rst
ds_betaseries_file/
_inputs.pklz
result_ds_betaseries_file.pklz
_0x28d42fc7961a0aca8c79fda9548df19c.json
_node.pklz
_report/
report.rst
mapflow/
_ds_betaseries_file0/
result__ds_betaseries_file0.pklz
_inputs.pklz
_0x44381d2af49bd9a22d139b0e53a1fa76.json
_node.pklz
_report/
report.rst
_ds_betaseries_file1/
_inputs.pklz
_node.pklz
_0x01f582a2d3dd0285859e8f67d9f6bd32.json
result__ds_betaseries_file1.pklz
_report/
report.rst
_ds_betaseries_file2/
_0xe7f43322abca06bcb89a9b07c69551b9.json
_inputs.pklz
result__ds_betaseries_file2.pklz
_node.pklz
_report/
report.rst
ds_correlation_matrix/
_inputs.pklz
_node.pklz
result_ds_correlation_matrix.pklz
_0x66b350ac65bb05172d17060a0e5094e1.json
_report/
report.rst
mapflow/
_ds_correlation_matrix1/
_inputs.pklz
_0xc309e1f1ead84d778882848909ec9cc0.json
_node.pklz
result__ds_correlation_matrix1.pklz
_report/
report.rst
_ds_correlation_matrix2/
_inputs.pklz
_0xf457a82159225d095e02b2d96045c7b7.json
result__ds_correlation_matrix2.pklz
_node.pklz
_report/
report.rst
_ds_correlation_matrix0/
_inputs.pklz
_0xc90f166f9433c553be90f25b1ed29a37.json
result__ds_correlation_matrix0.pklz
_node.pklz
_report/
report.rst
ds_correlation_fig/
_inputs.pklz
_0x3148251960ac02cbbd95e87293ded94e.json
_node.pklz
result_ds_correlation_fig.pklz
_report/
report.rst
mapflow/
_ds_correlation_fig2/
_inputs.pklz
_0x035e99cdf7cf49e86ce7c9d93244592b.json
_node.pklz
result__ds_correlation_fig2.pklz
_report/
report.rst
_ds_correlation_fig0/
_inputs.pklz
result__ds_correlation_fig0.pklz
_node.pklz
_0xa7ee3053a75350f0701acdaeb61e2b0c.json
_report/
report.rst
_ds_correlation_fig1/
_0x8bccd74bfe951cb0e469972ee708061d.json
_inputs.pklz
result__ds_correlation_fig1.pklz
_node.pklz
_report/
report.rst
betaseries_wf/
05f89e1eb2a40b75543a0ece936c065ae586c757/
betaseries_node/
desc-congruent_betaseries.nii.gz
_inputs.pklz
_0x0169097708d3cb942b1eaa2035ccf304.json
desc-residuals_bold.nii.gz
desc-congruent_betaseries_censored.nii.gz
_node.pklz
desc-incongruent_betaseries_censored.nii.gz
desc-neutral_betaseries.nii.gz
desc-neutral_betaseries_censored.nii.gz
desc-incongruent_betaseries.nii.gz
result_betaseries_node.pklz
_report/
report.rst
nibetaseries/
logs/
CITATION.md
CITATION.bib
CITATION.tex
sub-001/
func/
sub-001_task-stroop_space-MNI152NLin2009cAsym_desc-congruent_correlation.tsv
sub-001_task-stroop_space-MNI152NLin2009cAsym_desc-congruent_betaseries.nii.gz
sub-001_task-stroop_space-MNI152NLin2009cAsym_desc-congruent_correlation.svg
sub-001_task-stroop_space-MNI152NLin2009cAsym_desc-incongruent_correlation.svg
sub-001_task-stroop_space-MNI152NLin2009cAsym_desc-incongruent_betaseries.nii.gz
sub-001_task-stroop_space-MNI152NLin2009cAsym_desc-neutral_correlation.svg
sub-001_task-stroop_space-MNI152NLin2009cAsym_desc-incongruent_correlation.tsv
sub-001_task-stroop_space-MNI152NLin2009cAsym_desc-neutral_correlation.tsv
sub-001_task-stroop_space-MNI152NLin2009cAsym_desc-neutral_betaseries.nii.gz
fmriprep/
fMRIPrep.sqlite
dataset_description.json
sub-001/
func/
sub-001_task-stroop_desc-confounds_regressors.tsv
sub-001_task-stroop_space-MNI152NLin2009cAsym_desc-brain_mask.nii.gz
sub-001_task-stroop_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
data/
Schaefer2018_100Parcels_7Networks_order.tsv
Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz
Schaefer2018_100Parcels_7Networks_order.txt
sub-001/
anat/
sub-001_T1w.nii.gz
func/
sub-001_task-stroop_bold.nii.gz
sub-001_task-stroop_events.tsv
Collect correlation results¶
output_path = os.path.join(out_dir, "nibetaseries", "sub-001", "func")
trial_types = ['congruent', 'incongruent', 'neutral']
filename_template = ('sub-001_task-stroop_space-MNI152NLin2009cAsym_'
'desc-{trial_type}_{suffix}.{ext}')
pd_dict = {}
for trial_type in trial_types:
fname = filename_template.format(trial_type=trial_type, suffix='correlation', ext='tsv')
file_path = os.path.join(output_path, fname)
pd_dict[trial_type] = pd.read_csv(file_path, sep='\t', na_values="n/a", index_col=0)
# display example matrix
print(pd_dict[trial_type].head())
Out:
LH_Vis_1 LH_Vis_2 ... RH_Default_PCC_1 RH_Default_PCC_2
LH_Vis_1 NaN 0.218441 ... -0.229994 0.030440
LH_Vis_2 0.218441 NaN ... -0.272071 0.051983
LH_Vis_3 -0.193264 0.448418 ... 0.087879 0.083368
LH_Vis_4 0.122960 0.526585 ... -0.185481 0.188477
LH_Vis_5 0.284347 0.534536 ... -0.393536 -0.288098
[5 rows x 100 columns]
Graph the correlation results¶
fig, axes = plt.subplots(nrows=3, ncols=1, sharex=True, sharey=True, figsize=(10, 30),
gridspec_kw={'wspace': 0.025, 'hspace': 0.075})
cbar_ax = fig.add_axes([.91, .3, .03, .4])
r = 0
for trial_type, df in pd_dict.items():
g = sns.heatmap(df, ax=axes[r], vmin=-.5, vmax=1., square=True,
cbar=True, cbar_ax=cbar_ax)
axes[r].set_title(trial_type)
# iterate over rows
r += 1
plt.tight_layout()
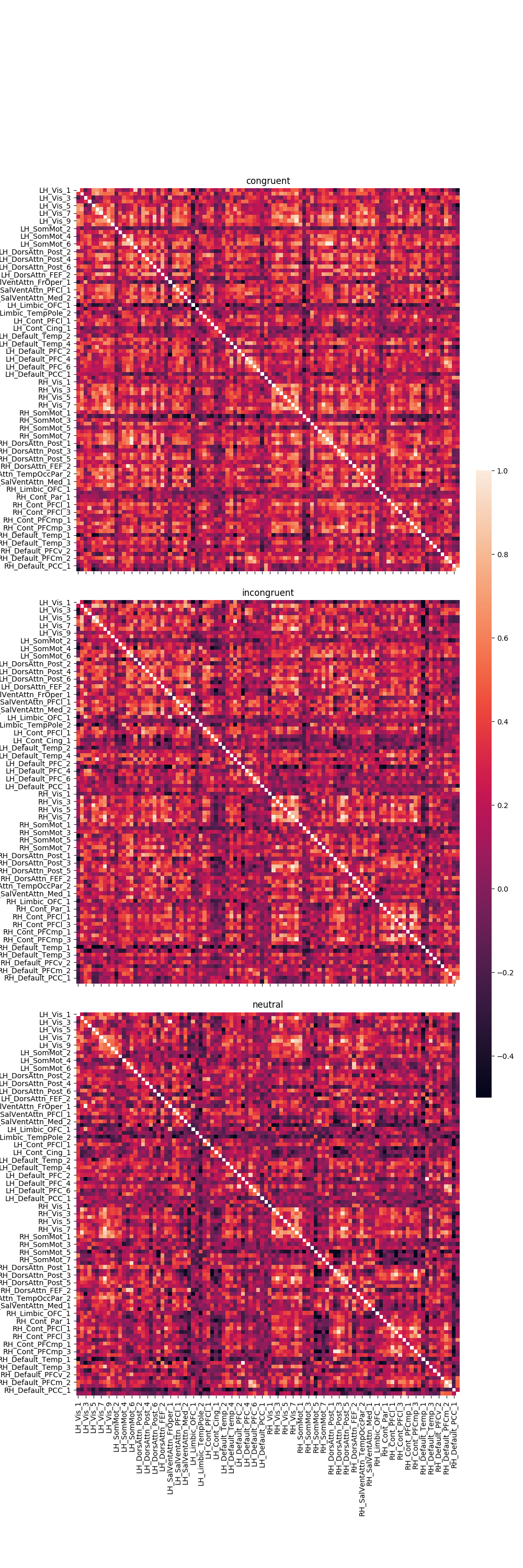
Out:
/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.6.0/lib/python3.7/site-packages/matplotlib/figure.py:2299: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
warnings.warn("This figure includes Axes that are not compatible "
Collect beta map results¶
nii_dict = {}
for trial_type in trial_types:
fname = filename_template.format(trial_type=trial_type, suffix='betaseries', ext='nii.gz')
file_path = os.path.join(output_path, fname)
nii_dict[trial_type] = nib.load(file_path)
# view incongruent beta_maps
nib.viewers.OrthoSlicer3D(nii_dict['incongruent'].get_fdata(),
title="Incongruent Betas").set_position(10, 13, 10)
Graph beta map standard deviation¶
We can find where the betas have the highest standard deviation for each trial type. Unsuprisingly, the largest deviations are near the edge of the brain mask and the subcortical regions.
# standard deviations for each trial type
std_dict = {tt: nib.Nifti1Image(img.get_fdata().std(axis=-1), img.affine, img.header)
for tt, img in nii_dict.items()}
fig, axes = plt.subplots(nrows=3, ncols=1, figsize=(10, 20))
for idx, (trial_type, nii) in enumerate(std_dict.items()):
plotting.plot_stat_map(nii, title=trial_type, cut_coords=(0, 0, 0),
threshold=5, vmax=20, axes=axes[idx])
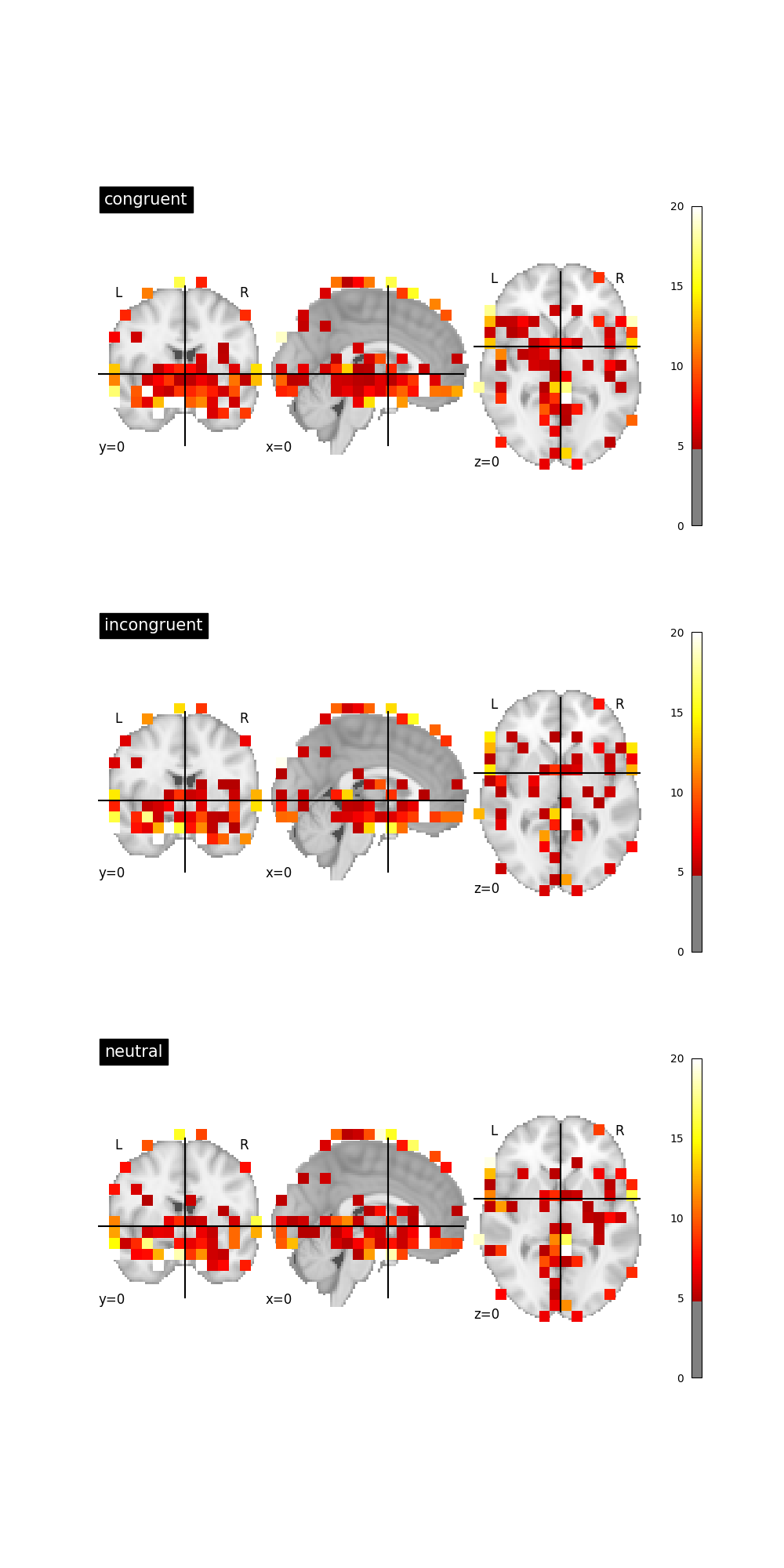
References¶
- notebook-1
Timothy D Verstynen. The organization and dynamics of corticostriatal pathways link the medial orbitofrontal cortex to future behavioral responses. Journal of Neurophysiology, 112(10):2457–2469, 2014. URL: https://doi.org/10.1152/jn.00221.2014, doi:10.1152/jn.00221.2014.
Total running time of the script: ( 1 minutes 29.186 seconds)