Note
Click here to download the full example code
Running NiBetaSeries using ds000164 (Stroop Task)¶
This example runs through a basic call of NiBetaSeries using
the commandline entry point nibs.
While this example is using python, typically nibs will be
called directly on the commandline.
Import all the necessary packages¶
import tempfile # make a temporary directory for files
import os # interact with the filesystem
import urllib.request # grad data from internet
import tarfile # extract files from tar
from subprocess import Popen, PIPE, STDOUT # enable calling commandline
import matplotlib.pyplot as plt # manipulate figures
import seaborn as sns # display results
import pandas as pd # manipulate tabular data
Download relevant data from ds000164 (and Atlas Files)¶
The subject data came from openneuro. The atlas data came from a recently published parcellation in a publically accessible github repository.
# atlas github repo for reference:
"""https://github.com/ThomasYeoLab/CBIG/raw/master/stable_projects/\
brain_parcellation/Schaefer2018_LocalGlobal/Parcellations/MNI/"""
data_dir = tempfile.mkdtemp()
print('Our working directory: {}'.format(data_dir))
# download the tar data
url = "https://www.dropbox.com/s/qoqbiya1ou7vi78/ds000164-test_v1.tar.gz?dl=1"
tar_file = os.path.join(data_dir, "ds000164.tar.gz")
u = urllib.request.urlopen(url)
data = u.read()
u.close()
# write tar data to file
with open(tar_file, "wb") as f:
f.write(data)
# extract the data
tar = tarfile.open(tar_file, mode='r|gz')
tar.extractall(path=data_dir)
os.remove(tar_file)
Out:
Our working directory: /tmp/tmpuu2igxzh
Display the minimal dataset necessary to run nibs¶
# https://stackoverflow.com/questions/9727673/list-directory-tree-structure-in-python
def list_files(startpath):
for root, dirs, files in os.walk(startpath):
level = root.replace(startpath, '').count(os.sep)
indent = ' ' * 4 * (level)
print('{}{}/'.format(indent, os.path.basename(root)))
subindent = ' ' * 4 * (level + 1)
for f in files:
print('{}{}'.format(subindent, f))
list_files(data_dir)
Out:
tmpuu2igxzh/
ds000164/
T1w.json
README
task-stroop_bold.json
dataset_description.json
task-stroop_events.json
CHANGES
derivatives/
data/
Schaefer2018_100Parcels_7Networks_order.txt
Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz
fmriprep/
sub-001/
func/
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_brainmask.nii.gz
sub-001_task-stroop_bold_confounds.tsv
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz
sub-001/
anat/
sub-001_T1w.nii.gz
func/
sub-001_task-stroop_bold.nii.gz
sub-001_task-stroop_events.tsv
Manipulate events file so it satifies assumptions¶
1. the correct column has 1’s and 0’s corresponding to correct and incorrect, respectively. 2. the condition column is renamed to trial_type nibs currently depends on the “correct” column being binary and the “trial_type” column to contain the trial types of interest.
read the file¶
events_file = os.path.join(data_dir,
"ds000164",
"sub-001",
"func",
"sub-001_task-stroop_events.tsv")
events_df = pd.read_csv(events_file, sep='\t', na_values="n/a")
print(events_df.head())
Out:
onset duration correct condition response_time
0 0.342 1 Y neutral 1.186
1 3.345 1 Y congruent 0.667
2 12.346 1 Y congruent 0.614
3 15.349 1 Y neutral 0.696
4 18.350 1 Y neutral 0.752
change the Y/N to 1/0¶
events_df['correct'].replace({"Y": 1, "N": 0}, inplace=True)
print(events_df.head())
Out:
onset duration correct condition response_time
0 0.342 1 1 neutral 1.186
1 3.345 1 1 congruent 0.667
2 12.346 1 1 congruent 0.614
3 15.349 1 1 neutral 0.696
4 18.350 1 1 neutral 0.752
replace condition with trial_type¶
events_df.rename({"condition": "trial_type"}, axis='columns', inplace=True)
print(events_df.head())
Out:
onset duration correct trial_type response_time
0 0.342 1 1 neutral 1.186
1 3.345 1 1 congruent 0.667
2 12.346 1 1 congruent 0.614
3 15.349 1 1 neutral 0.696
4 18.350 1 1 neutral 0.752
save the file¶
events_df.to_csv(events_file, sep="\t", na_rep="n/a", index=False)
Manipulate the region order file¶
There are several adjustments to the atlas file that need to be completed before we can pass it into nibs. Importantly, the relevant column names MUST be named “index” and “regions”. “index” refers to which integer within the file corresponds to which region in the atlas nifti file. “regions” refers the name of each region in the atlas nifti file.
read the atlas file¶
atlas_txt = os.path.join(data_dir,
"ds000164",
"derivatives",
"data",
"Schaefer2018_100Parcels_7Networks_order.txt")
atlas_df = pd.read_csv(atlas_txt, sep="\t", header=None)
print(atlas_df.head())
Out:
0 1 2 3 4 5
0 1 7Networks_LH_Vis_1 120 18 131 0
1 2 7Networks_LH_Vis_2 120 18 132 0
2 3 7Networks_LH_Vis_3 120 18 133 0
3 4 7Networks_LH_Vis_4 120 18 135 0
4 5 7Networks_LH_Vis_5 120 18 136 0
drop coordinate columns¶
atlas_df.drop([2, 3, 4, 5], axis='columns', inplace=True)
print(atlas_df.head())
Out:
0 1
0 1 7Networks_LH_Vis_1
1 2 7Networks_LH_Vis_2
2 3 7Networks_LH_Vis_3
3 4 7Networks_LH_Vis_4
4 5 7Networks_LH_Vis_5
rename columns with the approved headings: “index” and “regions”¶
atlas_df.rename({0: 'index', 1: 'regions'}, axis='columns', inplace=True)
print(atlas_df.head())
Out:
index regions
0 1 7Networks_LH_Vis_1
1 2 7Networks_LH_Vis_2
2 3 7Networks_LH_Vis_3
3 4 7Networks_LH_Vis_4
4 5 7Networks_LH_Vis_5
remove prefix “7Networks”¶
atlas_df.replace(regex={'7Networks_(.*)': '\\1'}, inplace=True)
print(atlas_df.head())
Out:
index regions
0 1 LH_Vis_1
1 2 LH_Vis_2
2 3 LH_Vis_3
3 4 LH_Vis_4
4 5 LH_Vis_5
write out the file as .tsv¶
atlas_tsv = atlas_txt.replace(".txt", ".tsv")
atlas_df.to_csv(atlas_tsv, sep="\t", index=False)
Run nibs¶
out_dir = os.path.join(data_dir, "ds000164", "derivatives")
work_dir = os.path.join(out_dir, "work")
atlas_mni_file = os.path.join(data_dir,
"ds000164",
"derivatives",
"data",
"Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz")
cmd = """\
nibs -c WhiteMatter CSF \
--participant_label 001 \
-w {work_dir} \
-a {atlas_mni_file} \
-l {atlas_tsv} \
{bids_dir} \
fmriprep \
{out_dir} \
participant
""".format(atlas_mni_file=atlas_mni_file,
atlas_tsv=atlas_tsv,
bids_dir=os.path.join(data_dir, "ds000164"),
out_dir=out_dir,
work_dir=work_dir)
# call nibs
p = Popen(cmd, shell=True, stdout=PIPE, stderr=STDOUT)
while True:
line = p.stdout.readline()
if not line:
break
print(line)
Out:
b'190129-14:37:59,784 nipype.workflow INFO:\n'
b"\t Workflow nibetaseries_participant_wf settings: ['check', 'execution', 'logging', 'monitoring']\n"
b'190129-14:37:59,795 nipype.workflow INFO:\n'
b'\t Running in parallel.\n'
b'190129-14:37:59,803 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b"/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/site-packages/grabbit/core.py:449: UserWarning: Domain with name 'bids' already exists; returning existing Domain configuration.\n"
b' warnings.warn(msg)\n'
b'190129-14:37:59,853 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.betaseries_wf.betaseries_node" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/betaseries_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/betaseries_node".\n'
b'190129-14:37:59,857 nipype.workflow INFO:\n'
b'\t [Node] Running "betaseries_node" ("nibetaseries.interfaces.nistats.BetaSeries")\n'
b'190129-14:38:01,808 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 1 tasks, and 0 jobs ready. Free memory (GB): 6.81/7.01, Free processors: 3/4.\n'
b' Currently running:\n'
b' * nibetaseries_participant_wf.single_subject001_wf.betaseries_wf.betaseries_node\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/importlib/_bootstrap.py:222: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88\n'
b' return f(*args, **kwds)\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/site-packages/nibabel/nifti1.py:582: DeprecationWarning: The binary mode of fromstring is deprecated, as it behaves surprisingly on unicode inputs. Use frombuffer instead\n'
b' ext_def = np.fromstring(ext_def, dtype=np.int32)\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b"/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/site-packages/nistats/hemodynamic_models.py:268: DeprecationWarning: object of type <class 'numpy.float64'> cannot be safely interpreted as an integer.\n"
b' frame_times.max() * (1 + 1. / (n - 1)), n_hr)\n'
b"/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/site-packages/nistats/hemodynamic_models.py:55: DeprecationWarning: object of type <class 'float'> cannot be safely interpreted as an integer.\n"
b' time_stamps = np.linspace(0, time_length, float(time_length) / dt)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'190129-14:40:06,940 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.betaseries_wf.betaseries_node".\n'
b'190129-14:40:07,946 nipype.workflow INFO:\n'
b'\t [Job 0] Completed (nibetaseries_participant_wf.single_subject001_wf.betaseries_wf.betaseries_node).\n'
b'190129-14:40:07,949 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190129-14:40:09,949 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190129-14:40:09,989 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node0" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/atlas_corr_node/mapflow/_atlas_corr_node0".\n'
b'190129-14:40:09,990 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node1" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/atlas_corr_node/mapflow/_atlas_corr_node1".\n'
b'190129-14:40:09,993 nipype.workflow INFO:\n'
b'\t [Node] Running "_atlas_corr_node0" ("nibetaseries.interfaces.nilearn.AtlasConnectivity")\n'
b'190129-14:40:09,994 nipype.workflow INFO:\n'
b'\t [Node] Running "_atlas_corr_node1" ("nibetaseries.interfaces.nilearn.AtlasConnectivity")\n'
b'190129-14:40:09,997 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node2" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/atlas_corr_node/mapflow/_atlas_corr_node2".\n'
b'190129-14:40:10,0 nipype.workflow INFO:\n'
b'\t [Node] Running "_atlas_corr_node2" ("nibetaseries.interfaces.nilearn.AtlasConnectivity")\n'
b'[NiftiLabelsMasker.fit_transform] loading data from /tmp/tmpuu2igxzh/ds000164/derivatives/data/Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz\n'
b'Resampling labels\n'
b'[NiftiLabelsMasker.transform_single_imgs] Loading data from /tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/betaseries_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/betaseries_node/betaseries_trialtyp\n'
b'[NiftiLabelsMasker.transform_single_imgs] Extracting region signals\n'
b'[NiftiLabelsMasker.transform_single_imgs] Cleaning extracted signals\n'
b'190129-14:40:10,909 nipype.workflow INFO:\n'
b'\t [Node] Finished "_atlas_corr_node1".\n'
b'[NiftiLabelsMasker.fit_transform] loading data from /tmp/tmpuu2igxzh/ds000164/derivatives/data/Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz\n'
b'Resampling labels\n'
b'[NiftiLabelsMasker.transform_single_imgs] Loading data from /tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/betaseries_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/betaseries_node/betaseries_trialtyp\n'
b'[NiftiLabelsMasker.transform_single_imgs] Extracting region signals\n'
b'[NiftiLabelsMasker.transform_single_imgs] Cleaning extracted signals\n'
b'190129-14:40:10,917 nipype.workflow INFO:\n'
b'\t [Node] Finished "_atlas_corr_node0".\n'
b'[NiftiLabelsMasker.fit_transform] loading data from /tmp/tmpuu2igxzh/ds000164/derivatives/data/Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz\n'
b'Resampling labels\n'
b'[NiftiLabelsMasker.transform_single_imgs] Loading data from /tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/betaseries_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/betaseries_node/betaseries_trialtyp\n'
b'[NiftiLabelsMasker.transform_single_imgs] Extracting region signals\n'
b'[NiftiLabelsMasker.transform_single_imgs] Cleaning extracted signals\n'
b'190129-14:40:10,919 nipype.workflow INFO:\n'
b'\t [Node] Finished "_atlas_corr_node2".\n'
b'190129-14:40:11,951 nipype.workflow INFO:\n'
b'\t [Job 4] Completed (_atlas_corr_node0).\n'
b'190129-14:40:11,952 nipype.workflow INFO:\n'
b'\t [Job 5] Completed (_atlas_corr_node1).\n'
b'190129-14:40:11,952 nipype.workflow INFO:\n'
b'\t [Job 6] Completed (_atlas_corr_node2).\n'
b'190129-14:40:11,954 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190129-14:40:12,0 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.correlation_wf.atlas_corr_node" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/atlas_corr_node".\n'
b'190129-14:40:12,3 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node0" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/atlas_corr_node/mapflow/_atlas_corr_node0".\n'
b'190129-14:40:12,4 nipype.workflow INFO:\n'
b'\t [Node] Cached "_atlas_corr_node0" - collecting precomputed outputs\n'
b'190129-14:40:12,4 nipype.workflow INFO:\n'
b'\t [Node] "_atlas_corr_node0" found cached.\n'
b'190129-14:40:12,5 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node1" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/atlas_corr_node/mapflow/_atlas_corr_node1".\n'
b'190129-14:40:12,6 nipype.workflow INFO:\n'
b'\t [Node] Cached "_atlas_corr_node1" - collecting precomputed outputs\n'
b'190129-14:40:12,6 nipype.workflow INFO:\n'
b'\t [Node] "_atlas_corr_node1" found cached.\n'
b'190129-14:40:12,7 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_atlas_corr_node2" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/atlas_corr_node/mapflow/_atlas_corr_node2".\n'
b'190129-14:40:12,8 nipype.workflow INFO:\n'
b'\t [Node] Cached "_atlas_corr_node2" - collecting precomputed outputs\n'
b'190129-14:40:12,8 nipype.workflow INFO:\n'
b'\t [Node] "_atlas_corr_node2" found cached.\n'
b'190129-14:40:12,10 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.correlation_wf.atlas_corr_node".\n'
b'190129-14:40:13,952 nipype.workflow INFO:\n'
b'\t [Job 1] Completed (nibetaseries_participant_wf.single_subject001_wf.correlation_wf.atlas_corr_node).\n'
b'190129-14:40:13,954 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190129-14:40:15,955 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190129-14:40:15,995 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_rename_matrix_node0" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/rename_matrix_node/mapflow/_rename_matrix_node0".\n'
b'190129-14:40:15,996 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_rename_matrix_node1" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/rename_matrix_node/mapflow/_rename_matrix_node1".\n'
b'190129-14:40:15,997 nipype.workflow INFO:\n'
b'\t [Node] Running "_rename_matrix_node0" ("nipype.interfaces.utility.wrappers.Function")\n'
b'190129-14:40:15,998 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_rename_matrix_node2" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/rename_matrix_node/mapflow/_rename_matrix_node2".\n'
b'190129-14:40:15,999 nipype.workflow INFO:\n'
b'\t [Node] Running "_rename_matrix_node1" ("nipype.interfaces.utility.wrappers.Function")\n'
b'190129-14:40:16,0 nipype.workflow INFO:\n'
b'\t [Node] Running "_rename_matrix_node2" ("nipype.interfaces.utility.wrappers.Function")\n'
b'190129-14:40:16,2 nipype.workflow INFO:\n'
b'\t [Node] Finished "_rename_matrix_node0".\n'
b'190129-14:40:16,3 nipype.workflow INFO:\n'
b'\t [Node] Finished "_rename_matrix_node1".\n'
b'190129-14:40:16,5 nipype.workflow INFO:\n'
b'\t [Node] Finished "_rename_matrix_node2".\n'
b'190129-14:40:17,956 nipype.workflow INFO:\n'
b'\t [Job 7] Completed (_rename_matrix_node0).\n'
b'190129-14:40:17,957 nipype.workflow INFO:\n'
b'\t [Job 8] Completed (_rename_matrix_node1).\n'
b'190129-14:40:17,957 nipype.workflow INFO:\n'
b'\t [Job 9] Completed (_rename_matrix_node2).\n'
b'190129-14:40:17,959 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190129-14:40:17,999 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.correlation_wf.rename_matrix_node" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/rename_matrix_node".\n'
b'190129-14:40:18,2 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_rename_matrix_node0" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/rename_matrix_node/mapflow/_rename_matrix_node0".\n'
b'190129-14:40:18,3 nipype.workflow INFO:\n'
b'\t [Node] Cached "_rename_matrix_node0" - collecting precomputed outputs\n'
b'190129-14:40:18,3 nipype.workflow INFO:\n'
b'\t [Node] "_rename_matrix_node0" found cached.\n'
b'190129-14:40:18,4 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_rename_matrix_node1" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/rename_matrix_node/mapflow/_rename_matrix_node1".\n'
b'190129-14:40:18,5 nipype.workflow INFO:\n'
b'\t [Node] Cached "_rename_matrix_node1" - collecting precomputed outputs\n'
b'190129-14:40:18,5 nipype.workflow INFO:\n'
b'\t [Node] "_rename_matrix_node1" found cached.\n'
b'190129-14:40:18,6 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_rename_matrix_node2" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/correlation_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/rename_matrix_node/mapflow/_rename_matrix_node2".\n'
b'190129-14:40:18,7 nipype.workflow INFO:\n'
b'\t [Node] Cached "_rename_matrix_node2" - collecting precomputed outputs\n'
b'190129-14:40:18,7 nipype.workflow INFO:\n'
b'\t [Node] "_rename_matrix_node2" found cached.\n'
b'190129-14:40:18,9 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.correlation_wf.rename_matrix_node".\n'
b'190129-14:40:19,958 nipype.workflow INFO:\n'
b'\t [Job 2] Completed (nibetaseries_participant_wf.single_subject001_wf.correlation_wf.rename_matrix_node).\n'
b'190129-14:40:19,960 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190129-14:40:21,961 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 3 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190129-14:40:22,6 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix0" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/ds_correlation_matrix/mapflow/_ds_correlation_matrix0".\n'
b'190129-14:40:22,7 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix1" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/ds_correlation_matrix/mapflow/_ds_correlation_matrix1".\n'
b'190129-14:40:22,8 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix0" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190129-14:40:22,9 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix1" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190129-14:40:22,12 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix0".\n'
b'190129-14:40:22,15 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix1".\n'
b'190129-14:40:22,16 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix2" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/ds_correlation_matrix/mapflow/_ds_correlation_matrix2".\n'
b'190129-14:40:22,17 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix2" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190129-14:40:22,21 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix2".\n'
b'190129-14:40:23,962 nipype.workflow INFO:\n'
b'\t [Job 10] Completed (_ds_correlation_matrix0).\n'
b'190129-14:40:23,966 nipype.workflow INFO:\n'
b'\t [Job 11] Completed (_ds_correlation_matrix1).\n'
b'190129-14:40:23,968 nipype.workflow INFO:\n'
b'\t [Job 12] Completed (_ds_correlation_matrix2).\n'
b'190129-14:40:23,970 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'190129-14:40:24,8 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "nibetaseries_participant_wf.single_subject001_wf.ds_correlation_matrix" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/ds_correlation_matrix".\n'
b'190129-14:40:24,12 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix0" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/ds_correlation_matrix/mapflow/_ds_correlation_matrix0".\n'
b'190129-14:40:24,19 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix0" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190129-14:40:24,22 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix0".\n'
b'190129-14:40:24,23 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix1" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/ds_correlation_matrix/mapflow/_ds_correlation_matrix1".\n'
b'190129-14:40:24,26 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix1" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190129-14:40:24,30 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix1".\n'
b'190129-14:40:24,31 nipype.workflow INFO:\n'
b'\t [Node] Setting-up "_ds_correlation_matrix2" in "/tmp/tmpuu2igxzh/ds000164/derivatives/work/NiBetaSeries_work/nibetaseries_participant_wf/single_subject001_wf/a7dc49b58761b73b7aeeeee9d35703cc8ab43895/ds_correlation_matrix/mapflow/_ds_correlation_matrix2".\n'
b'190129-14:40:24,34 nipype.workflow INFO:\n'
b'\t [Node] Running "_ds_correlation_matrix2" ("nibetaseries.interfaces.bids.DerivativesDataSink")\n'
b'190129-14:40:24,38 nipype.workflow INFO:\n'
b'\t [Node] Finished "_ds_correlation_matrix2".\n'
b'190129-14:40:24,40 nipype.workflow INFO:\n'
b'\t [Node] Finished "nibetaseries_participant_wf.single_subject001_wf.ds_correlation_matrix".\n'
b'190129-14:40:25,964 nipype.workflow INFO:\n'
b'\t [Job 3] Completed (nibetaseries_participant_wf.single_subject001_wf.ds_correlation_matrix).\n'
b'190129-14:40:25,966 nipype.workflow INFO:\n'
b'\t [MultiProc] Running 0 tasks, and 0 jobs ready. Free memory (GB): 7.01/7.01, Free processors: 4/4.\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/importlib/_bootstrap.py:222: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88\n'
b' return f(*args, **kwds)\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/importlib/_bootstrap.py:222: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88\n'
b' return f(*args, **kwds)\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/importlib/_bootstrap.py:222: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88\n'
b' return f(*args, **kwds)\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/site-packages/nipype/pipeline/engine/utils.py:307: DeprecationWarning: use "HasTraits.trait_set" instead\n'
b' result.outputs.set(**modify_paths(tosave, relative=True, basedir=cwd))\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 2 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 2 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 1 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'Computing run 1 out of 1 runs (go take a coffee, a big one)\n'
b'\n'
b'Computation of 1 runs done in 0 seconds\n'
b'\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/site-packages/nipype/pipeline/engine/utils.py:307: DeprecationWarning: use "HasTraits.trait_set" instead\n'
b' result.outputs.set(**modify_paths(tosave, relative=True, basedir=cwd))\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/importlib/_bootstrap.py:222: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88\n'
b' return f(*args, **kwds)\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/importlib/_bootstrap.py:222: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88\n'
b' return f(*args, **kwds)\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/importlib/_bootstrap.py:222: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88\n'
b' return f(*args, **kwds)\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/site-packages/nipype/pipeline/engine/utils.py:307: DeprecationWarning: use "HasTraits.trait_set" instead\n'
b' result.outputs.set(**modify_paths(tosave, relative=True, basedir=cwd))\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/importlib/_bootstrap.py:222: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88\n'
b' return f(*args, **kwds)\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/importlib/_bootstrap.py:222: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88\n'
b' return f(*args, **kwds)\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/importlib/_bootstrap.py:222: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88\n'
b' return f(*args, **kwds)\n'
b'/home/docs/checkouts/readthedocs.org/user_builds/nibetaseries/envs/v0.2.3/lib/python3.5/site-packages/nipype/pipeline/engine/utils.py:307: DeprecationWarning: use "HasTraits.trait_set" instead\n'
b' result.outputs.set(**modify_paths(tosave, relative=True, basedir=cwd))\n'
Observe generated outputs¶
list_files(data_dir)
Out:
tmpuu2igxzh/
ds000164/
T1w.json
README
task-stroop_bold.json
dataset_description.json
task-stroop_events.json
CHANGES
derivatives/
NiBetaSeries/
nibetaseries/
sub-001/
func/
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc_trialtype-congruent_matrix.tsv
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc_trialtype-incongruent_matrix.tsv
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc_trialtype-neutral_matrix.tsv
logs/
data/
Schaefer2018_100Parcels_7Networks_order.tsv
Schaefer2018_100Parcels_7Networks_order.txt
Schaefer2018_100Parcels_7Networks_order_FSLMNI152_2mm.nii.gz
work/
NiBetaSeries_work/
nibetaseries_participant_wf/
graph1.json
d3.js
index.html
graph.json
single_subject001_wf/
a7dc49b58761b73b7aeeeee9d35703cc8ab43895/
ds_correlation_matrix/
_0x71f4d002e2c91ce513522bea20d623bd.json
_node.pklz
result_ds_correlation_matrix.pklz
_inputs.pklz
_report/
report.rst
mapflow/
_ds_correlation_matrix0/
_node.pklz
result__ds_correlation_matrix0.pklz
_inputs.pklz
_0x88f8d315f589eeaac7d70cb68d8a014e.json
_report/
report.rst
_ds_correlation_matrix1/
_node.pklz
_inputs.pklz
_0x3f0a8f030067a681159da302b0d8678b.json
result__ds_correlation_matrix1.pklz
_report/
report.rst
_ds_correlation_matrix2/
_node.pklz
_inputs.pklz
_0xccb9f070c4554dd0dd86356895be3dff.json
result__ds_correlation_matrix2.pklz
_report/
report.rst
correlation_wf/
a7dc49b58761b73b7aeeeee9d35703cc8ab43895/
atlas_corr_node/
_node.pklz
result_atlas_corr_node.pklz
_inputs.pklz
_0xe429a02dadec120752df8309cfec5434.json
_report/
report.rst
mapflow/
_atlas_corr_node2/
fisher_z_correlation.tsv
_node.pklz
_inputs.pklz
_0xf833ae8eaec6073d5d1f2f9f3d78038b.json
result__atlas_corr_node2.pklz
_report/
report.rst
_atlas_corr_node0/
fisher_z_correlation.tsv
_node.pklz
_inputs.pklz
result__atlas_corr_node0.pklz
_0x0027e94ede3403f7800288af1d5de310.json
_report/
report.rst
_atlas_corr_node1/
fisher_z_correlation.tsv
_node.pklz
_inputs.pklz
_0x9e7d17fcd6da69a6071a10aa34bfd767.json
result__atlas_corr_node1.pklz
_report/
report.rst
rename_matrix_node/
_node.pklz
result_rename_matrix_node.pklz
_inputs.pklz
_0x5e36fb501fbee11912d6c173ac4f0573.json
_report/
report.rst
mapflow/
_rename_matrix_node1/
_node.pklz
_inputs.pklz
result__rename_matrix_node1.pklz
correlation-matrix_trialtype-incongruent.tsv
_0x09f938ebbd08bd01b0fdb9735da975cf.json
_report/
report.rst
_rename_matrix_node2/
_node.pklz
_inputs.pklz
result__rename_matrix_node2.pklz
correlation-matrix_trialtype-neutral.tsv
_0x09b2b427be3a30edca3d550c457949d7.json
_report/
report.rst
_rename_matrix_node0/
_node.pklz
_inputs.pklz
result__rename_matrix_node0.pklz
_0xab741b49debbe7844f004f5dabf45fd8.json
correlation-matrix_trialtype-congruent.tsv
_report/
report.rst
betaseries_wf/
a7dc49b58761b73b7aeeeee9d35703cc8ab43895/
betaseries_node/
_node.pklz
_inputs.pklz
betaseries_trialtype-neutral.nii.gz
betaseries_trialtype-congruent.nii.gz
betaseries_trialtype-incongruent.nii.gz
result_betaseries_node.pklz
_0xaeb6e1e256ee0af78f95368119ba2faa.json
_report/
report.rst
fmriprep/
sub-001/
func/
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_brainmask.nii.gz
sub-001_task-stroop_bold_confounds.tsv
sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz
sub-001/
anat/
sub-001_T1w.nii.gz
func/
sub-001_task-stroop_bold.nii.gz
sub-001_task-stroop_events.tsv
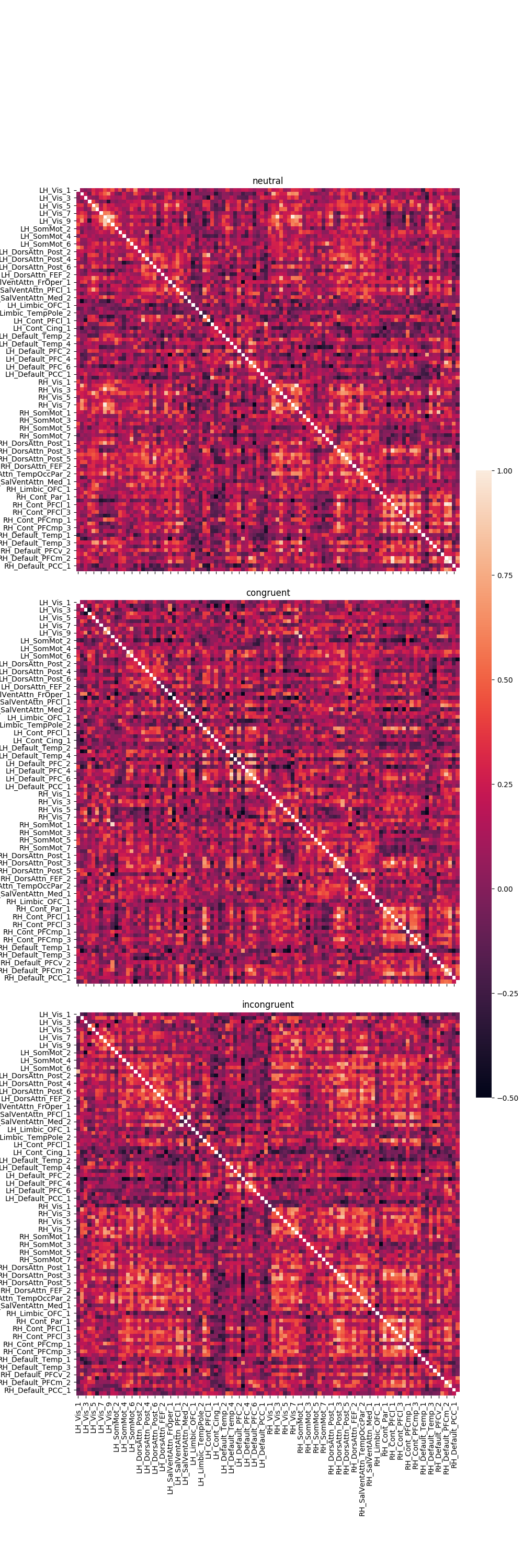
Collect results¶
corr_mat_path = os.path.join(out_dir, "NiBetaSeries", "nibetaseries", "sub-001", "func")
trial_types = ['congruent', 'incongruent', 'neutral']
filename_template = "sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc_trialtype-{trial_type}_matrix.tsv"
pd_dict = {}
for trial_type in trial_types:
file_path = os.path.join(corr_mat_path, filename_template.format(trial_type=trial_type))
pd_dict[trial_type] = pd.read_csv(file_path, sep='\t', na_values="n/a", index_col=0)
# display example matrix
print(pd_dict[trial_type].head())
Out:
LH_Vis_1 LH_Vis_2 ... RH_Default_PCC_1 RH_Default_PCC_2
LH_Vis_1 NaN 0.092135 ... 0.095624 0.016799
LH_Vis_2 0.092135 NaN ... -0.119613 -0.007679
LH_Vis_3 -0.003990 0.216346 ... 0.202673 0.177828
LH_Vis_4 0.075498 -0.088788 ... -0.019256 -0.034034
LH_Vis_5 0.314494 0.354525 ... -0.235334 0.032317
[5 rows x 100 columns]
Graph the results¶
fig, axes = plt.subplots(nrows=3, ncols=1, sharex=True, sharey=True, figsize=(10, 30),
gridspec_kw={'wspace': 0.025, 'hspace': 0.075})
cbar_ax = fig.add_axes([.91, .3, .03, .4])
r = 0
for trial_type, df in pd_dict.items():
g = sns.heatmap(df, ax=axes[r], vmin=-.5, vmax=1., square=True,
cbar=True, cbar_ax=cbar_ax)
axes[r].set_title(trial_type)
# iterate over rows
r += 1
plt.tight_layout()
Total running time of the script: ( 2 minutes 34.309 seconds)